Simplifying molecular cloning
Molecular cloning techniques usually involve multiple enzymatic steps, consisting of separate restriction digestion, dephosphorylation, and enzymatic ligation reactions. Alternating between the various enzymes and buffer conditions often requires purification steps, such as gel and column purification, or ethanol precipitation. These multiple steps add time and complexity to the protocol, and often result in the loss of DNA. Planning for restriction digest cloning of multiple fragments can also be time consuming, and designing larger restriction assemblies has additional challenges as larger constructs will have fewer unique restriction sites.
As with any molecular technique, it is beneficial to reach the desired goal in as few molecular cloning steps as possible. This limits potential errors due to repeated handling of samples and reagents, and avoids multiple experimental setups.
What is seamless cloning?
The seamless cloning method, also often called Gibson assembly, simplifies the process for molecular cloning of synthesized DNA molecules. This method makes it possible to include larger, more complex assemblies than traditional cloning methods.
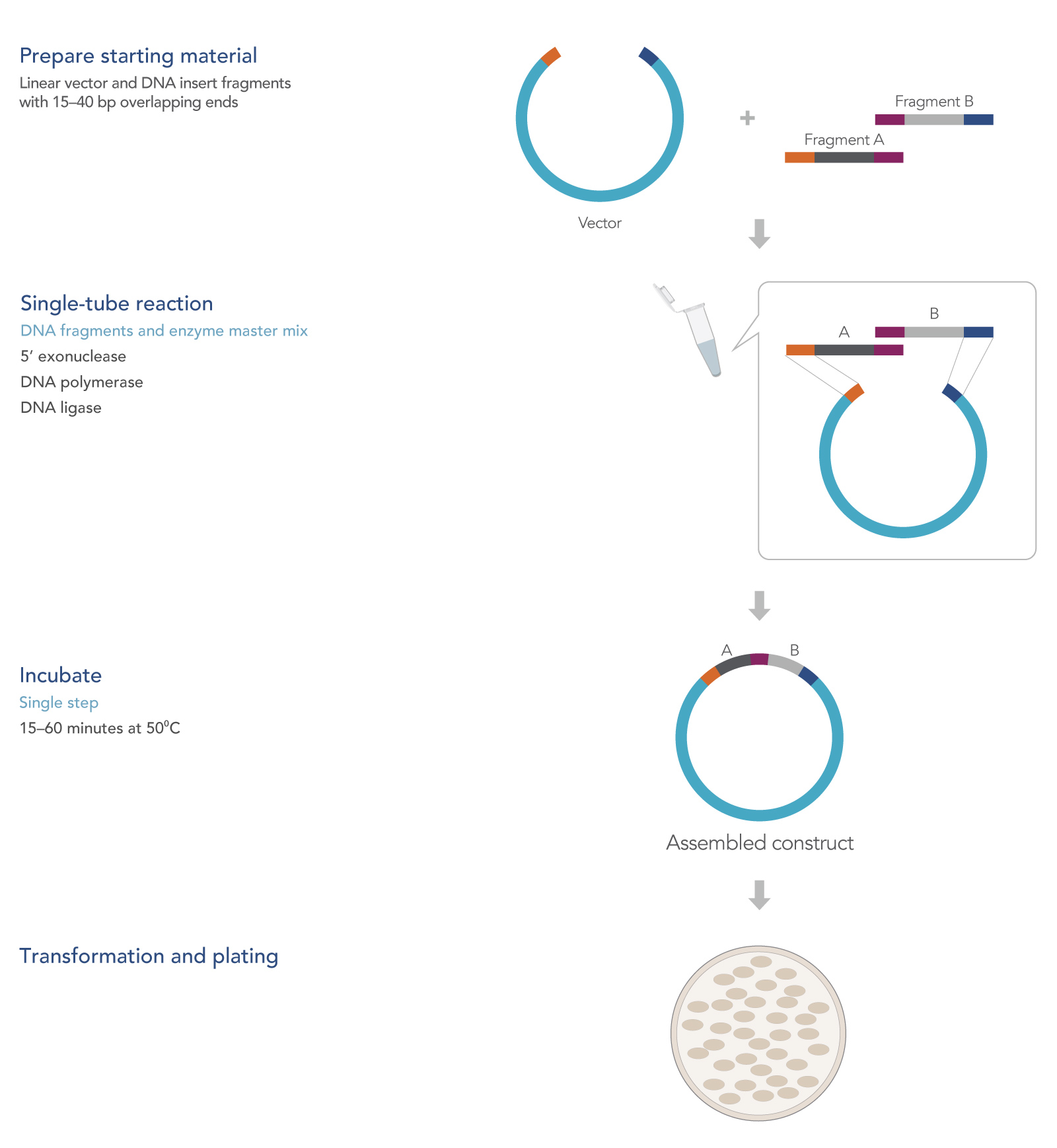
How seamless cloning works
In a single reaction, seamless cloning combines several overlapping DNA fragments to produce a ligated plasmid ready for transformation. The method relies on use of an enzyme mixture consisting of the mesophilic 5’ T5 exonuclease, a thermophilic DNA ligase, and a thermophilic proofreading DNA polymerase. Details of what happens in the reaction are outlined in Figure 1.
When added to DNA and placed at high temperatures, the exonuclease digests the double-stranded DNA from its 5’ ends. This results in 3’ single-stranded ends, which are then available to reanneal with complementary 3’ strands from an adjacent fragment.
For effective assembly and ligation, the ends of the DNA sequences are designed to overlap by 20–40 bases. The 5’ exonuclease activity exposes the overlapping bases, leaving variable length overhangs at the 3’ ends. These large overlaps are beneficial for joining fragments as they allow for highly specific hybridization of DNA fragments—compared to a typical restriction cloning reaction in which complementary overhangs are no more than 4 bases—at the restriction site.
Because the T5 exonuclease processes a random number of bases there will be single-stranded gaps between the hybridized 3’ ends and the remaining 5’ ends. The high-fidelity DNA polymerase fills the resulting gaps in a 5’-to-3’ direction, leaving only a single-stranded nick. The thermophilic DNA ligase then seals the nick and covalently links the fragments together.
Design considerations
A major benefit of the seamless cloning method is that it has fewer specific design requirements than other forms of cloning. For example, there is no need to look for endogenous recognition sites for restriction enzymes, or to insert such sites that could subsequently affect the usefulness of the final DNA construct. Therefore, the basic principles of design for this method are simple:
- For the seamless cloning to work the ends of the vector and the insert(s) must complement one another. In general, 30 bases of complementarity are optimal at each end. Ideally the ends should be free from secondary structure. If using a commercial kit, make sure to follow the manufacturer's instructions for the optimal length of homology arms.
- If you are planning to assemble DNA that has been produced from PCR, primers can be designed so that each primer has 15 bases of overlap into what will be the adjacent fragment to create the 30 bases of overlap.
- Since hybridization of long 3’ overhangs is similar to hybridization of PCR primers, avoid designing 3’ overlaps in regions with repeated DNA motifs or repeated bases that can hybridize though improperly aligned.
- If the final construct is to be linear rather than a fragment subcloned into a plasmid vector, the ends of the DNA will be retracted at each 5’ end. If desired, these ends can be filled in a separate reaction using a short oligonucleotide primer complementary to the 3’ end. This is not necessary for circular constructs, as all gaps are filled, and the ends ligated during the seamless cloning reaction.
To learn more about seamless cloning and other gene assembly methods, download the DNA Cloning Guide.


 Processing
Processing

