Phosphoramidite synthesis vs. post-synthesis conjugation
Modifications can be incorporated into synthetic oligonucleotides in a variety of ways. Standard DNA bases are synthetically coupled via phosphoramidite chemistry. The reaction proceeds in the 3' to 5' direction where the 5' hydroxyl group of each base attaches to the 3' phosphate group of the next base. Many modifications, such as 6-FAM, standard biotin, and internal Cy3, can be attached via phosphoramidite chemistry directly on the synthesis column (Figure 1).

Most modifications are attached using phosphoramidite chemistry. However, some modifications, such as NHS esters and click chemistry modifications, are attached post synthesis. The following sections will focus on the common post-synthesis conjugations performed at IDT.
NHS ester modifications
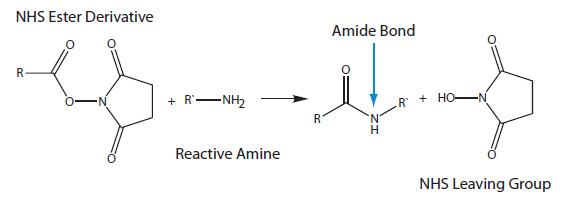
NHS ester modifications contain an NHS (N-hydroxysuccinimide) group that reacts with an amine group to form an amide (Figure 2).
Typically, 5' NHS esters are attached through an Amino Modifier C6 group, internal NHS esters through an Amino Modifier C6 dT, and 3' NHS esters via an amino group linked to the controlled pore glass (CPG) beads used as the synthesis support (Figure 3).When ordering modified oligos, certain modifications like fluorescent dyes may have to be attached to the oligo via the NHS ester attachment route. For example, 5' MAX™ or 5' JOE™ will be designated as 5' MAX (NHS Ester) or 5' JOE (NHS Ester) when ordered.

What is click chemistry?
The nature and mechanism of click chemistry was described by Dr K. B. Sharpless at the Scripps Institute in 2001 [1,2]. A click chemistry reaction entails coupling azide and alkyne groups through a copper-catalyzed reaction, forming a 1,2,3-triazole. This reaction is thermodynamically favorable, resulting in an irreversible reaction with no side products (Figure 4). There are also copper-free click reactions [3].

IDT offers a variety of oligo modifications that leave a free azide or alkyne group available for further click conjugation, giving researchers the freedom to conjugate molecules of their choice to create custom oligo modifications. Alternatively, IDT can do the click conjugation for you by including a reactive alkyne group in the oligonucleotide during synthesis. After synthesis, deprotection, and initial purification, the alkyne group is reacted with a modification containing an azide functional group.
All 5' click modifications conjugate through a 5' hexynyl group (Figure 5A) while internal click modifications conjugate through an internal alkyne (Figure 5B). The internal alkyne group is attached to dT, meaning any internal modification attached via click chemistry will incorporate an additional T base into the oligonucleotide sequence. To identify oligo modifications attached via click chemistry, look for (azide) after the modification name, such as 6-FAM (azide). IDT can also provide a 3' Alkyne Modifier as a non-catalog request (Figure 5C).

IDT NHS ester and click chemistry modifications
Table 1 shows current IDT catalog offerings of NHS ester and click chemistry modifications. Don't see the modification you need in our catalog? No worries. IDT routinely accepts requests for custom oligo modifications outside of our normal catalog offerings. Simply contact us to request non-catalog products or if you have questions about oligonucleotide modifications.
Table 1. NHS ester modifications.
| Modification name | 5' | Internal | 3' |
| 6-FAM | • | • | |
| Alexa Fluor® 488 | • | • | |
| Alexa Fluor 532 | • | • | |
| Alexa Fluor 546 | • | • | |
| Alexa Fluor 594 | • | • | |
| Alexa Fluor 647 | • | • | |
| Alexa Fluor 660 | • | • | |
| Alexa Fluor 750 | • | • | |
| ATTO™ 488 | • | • | |
| ATTO 532 | • | • | |
| ATTO 550 | • | • | |
| ATTO 565 | • | • | |
| ATTO Rho101 | • | • | |
| ATTO 590 | • | • | |
| ATTO 633 | • | • | |
| ATTO 647N | • | • | |
| Azide (NHS Ester) | • | • | • |
| Digoxigenin | • | • | |
| Dy 750™ | • | ||
| IRDye® 800 | • | ||
| JOE | • | • | |
| LightCycler® 640 | • | • | |
| MAX | • | • | |
| Rhodamine Green-X™ | • | • | |
| Rhodamine Red-X | • | • | |
| ROX | • | • | |
| TAMRA | • | • | • |
| Texas Red-X | • | • |


 Processing
Processing