Looking for CRISPR genome editing protocols? Read about our growing library of protocols and user methods—we may have what you need to get started. And, if you have a novel protocol for using Alt-R CRISPR RNA and/or nucleases, find out how to share it with the research community. <<CRISPR genome editing protocols and user methods>>
Challenges experienced with Jurkat cells
Derived from T lymphocytic cells, the Jurkat cell line has been cloned into numerous sublines that are used in research laboratories to study immune system signaling, disease impact on the immune system, susceptibility of cancers to drugs and radiation treatment, and viral infection through chemokine receptors (such as with HIV). However, these cells are known for being notoriously difficult to transfect. This presents a problem for researchers interested in applying CRISPR genome editing technologies in these cells.
Lipofection protocols are often used for adherent, immortalized, eukaryotic cell lines, with protocol specifics often requiring optimization for different cell lines. For primary cells, non-dividing cells, and difficult-to-transfect cells, such as Jurkat cells, another delivery method, such as electroporation, is often required. The goal is to identify conditions that confer maximal editing efficiency and minimal cell toxicity.
We have already presented an optimized lipofection protocol for CRISPR reagents in HEK293 cells (see Alt-R® CRISPR-Cas9 System Users Guide). This article describes the optimization of electroporation conditions for delivery of CRISPR RNAs and Cas9 nuclease into Jurkat cells (Clone E6-1), using the Neon® Transfection System (Thermo Fisher Scientific). We also present data showing successful genome editing using our identified electroporation conditions for this cell line. The approach can be applied to other difficult-to-transfect cells, such as B cells, macrophages, and monocytes.
Reagent format
Scientists at IDT have established that CRISPR RNAs and Cas9 protein are most effectively delivered to transfected cell lines as a ribonucleoprotein (RNP) complex.
As part of the IDT Alt-R CRISPR-Cas9 System reagents, we offer crRNA, tracrRNAs, and the S.p. Cas9 Nuclease 3NLS. The RNAs are length optimized and chemically modified to further enhance genome editing by rendering the oligos less prone to degradation by nucleases. These modified RNAs thus have greater stability that leads to higher editing efficiencies, especially when used with the S.p. Cas9 Nuclease 3NLS.
For our Jurkat cell delivery optimization experiments, we therefore chose to use RNPs consisting of modified CRISPR RNAs complexed with Cas9 nuclease.
Electroporation optimization
For the optimization experiments, we designed an Alt-R CRISPR-Cas9 System crRNA targeting the HPRT gene. The crRNA and tracrRNA were complexed in a 1:1 ratio with a final working concentration of 45 µM. We then generated RNP complexes by combining S.p. Cas9 Nuclease 3NLS protein and crRNA:tracrRNA in a 1:1.2 ratio, with a final working concentration of 18:21.6 µM.
The Jurkat cell line has been cloned into numerous sublines. For these experiments, we used Clone E6-1 Jurkat cells (ATCC® TIB-152™). 2 x 105 Jurkat cells were diluted in 10 µL Resuspension Buffer R (Neon Transfection System, Thermo Fisher Scientific) and added to 1 µL RNP complex. To test the effect of carrier DNA, 1 µL sequence optimized carrier DNA was added to a final concentration of 1.8 µM. For samples excluding carrier, we added 1 µL Buffer R.
Using a 10-µL Neon Transfection System tip, we followed the Neon® Optimization protocol (Publication Number MAN0001557, Revision A.0, page 22), which tests 24 electroporation settings, including voltage, pulse width, and number of pulses. Electroporated cells (10 µL) were mixed with 175 µL media, and 50 µL diluted cells were plated in triplicate in 100 µL pre-warmed media in a 96-well plate.
After 72 hours, we took images of the cell cultures to determine cell density. Genomic DNA was isolated to analyze genome editing using the T7EI mismatch endonuclease assay, as described in the Alt-R CRISPR-Cas9 System Users Guide. Note that the T7El assay underestimates total editing (see the sidebar, T7EI mismatch endonuclease assay for genome editing analysis)
Electroporation results
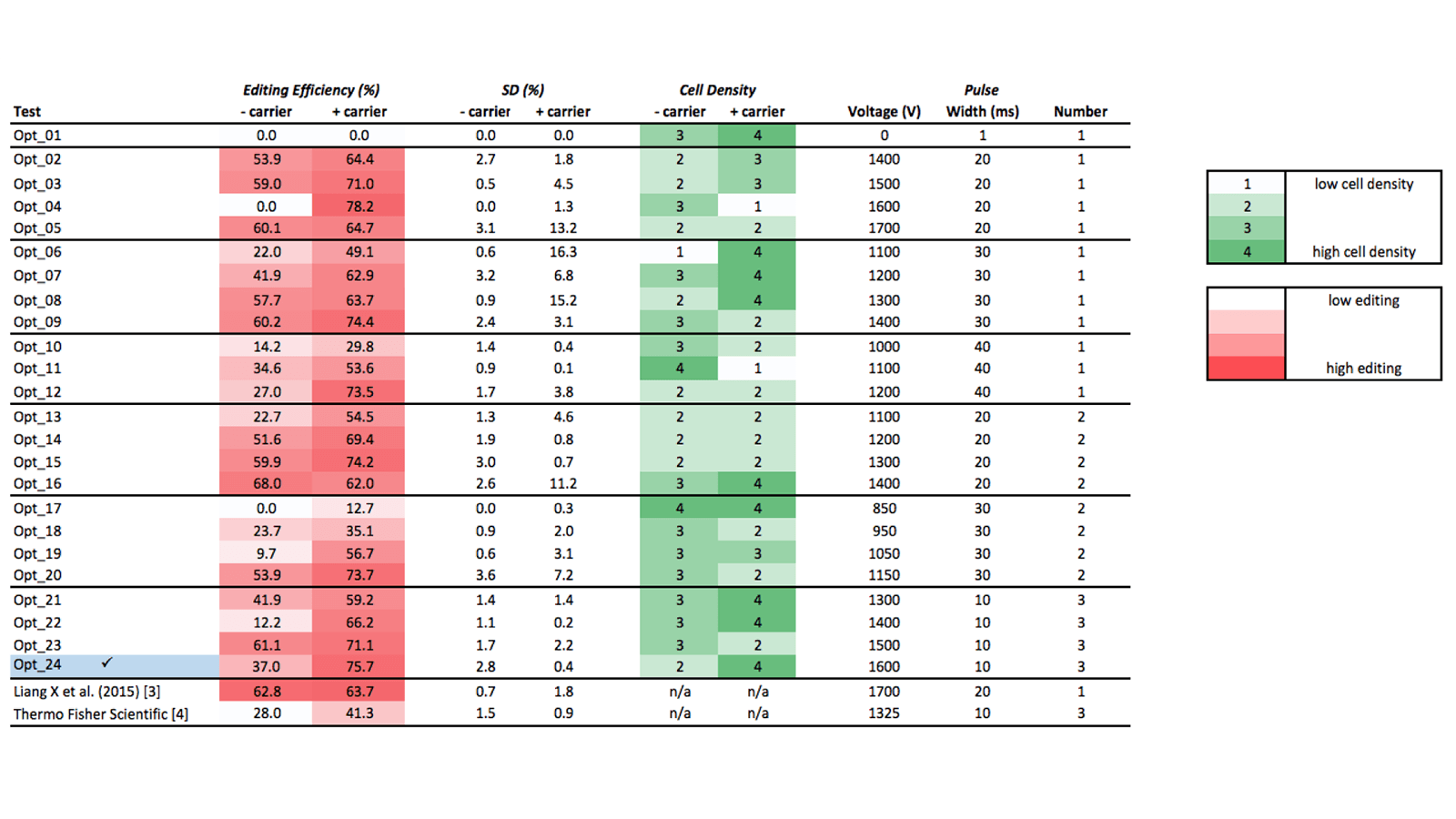
Table 1 lists the 24 electroporation test conditions. Cell density (reflective of cell toxicity) are scored using a 4 point scale; T7EI editing efficiency results are scored as percentages of detected edited alleles over non-edited alleles. Based on these two parameters, we identified optimal conditions for Neon electroporation of Jurkat cells as:
- Electroporate with 3 pulses of 10 milliseconds, at 1600 volts
- Include carrier DNA
Jurkat electroporation with CRISPR reagents leads to successful genome editing
The data presented here demonstrate that use of Alt-R® CRISPR-Cas9 System reagents for successful genome editing is not limited to easy-to-transfect cell types. With the resulting optimized electroporation conditions, we have shown Jurkat cell genome
modification >75% + 0.4%. Keep in mind that RNA, protein, or RNP delivery conditions will require optimization for each individual cell line.
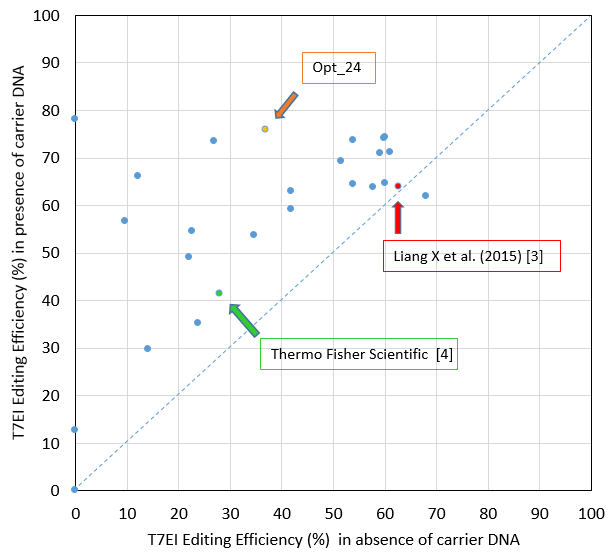
The editing efficiency we obtained in this study is substantially higher than that achieved with the Thermo Fisher Scientific protocol, as described on their website [4], and is also higher than that described by Liang et al [3] (Figure 2).
While these authors may have used distinct Jurkat subclones, which could explain some of the editing efficiency difference, our addition of a specific type of carrier DNA that enhances editing efficiency has clearly contributed to the superior
data we obtained.
In cell types where lipofection has proven ineffective, electroporation should be considered as an alternative RNP delivery method. The electroporation optimization experiment presented here can provide a starting point for optimization of CRISPR reagent delivery in other cell types.

