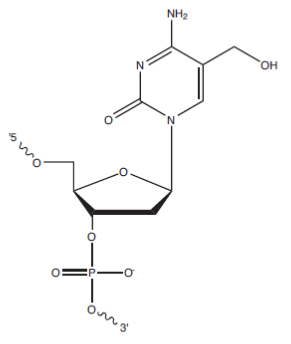
Quick facts:
Availability: DNA
Location:
5′, 3′, or internal
Scale: 100 nmol, 250 nmol, 1 µmol, 5 µmol, and 10 µmol
Purification: HPLC required
Ordering symbols:
For 5′ placement, use /55HydMe-dC/
For 3′ placement, use /35HydMe-dC/
For internal placement, use /i5HydMe-dC/
The methylation of deoxycytosine (dC) to 5-methylcytosine (5-mC) has long been studied for the important role it plays as an epigenetic marker in gene expression and regulation. Now researchers are turning their attention to 5-hydroxymethylcytosine (5-hmC), a recently discovered “sixth base” [1]. In mammalian cells, 5-hmC has been detected in all tissue types, with the highest levels observed in the brain and spinal cord [2,3]. Jin, Wu, et al. [4] were among the first to compare levels and locations of 5-hmC to 5-mC in human brain tissue, and reported its enrichment at promoters and in intragenic regions. It has been reported that 5-mC is converted to 5-hmC by TET proteins, which perform various roles in gene expression. While the exact biological function of 5-hmC is unknown, it has been hypothesized that it may be an intermediate in a demethylation pathway or may act as an additional epigenetic factor [5].
Ordering oligos with 5-Hydroxymethyl-dC
You can order oligos with this modification introduced at any position using standard. To order an oligonucleotide modified with 5-hydroxymethylcytosine: select 5-Hydroxymethyl-dC from the modifications tab on the oligonucleotide order page. Alternatively, use the appropriate modification code.
Other modifications to facilitate your research
See what other modifications are available from IDT. You can find a list of standard modifications we make to oligonucleotides on the Modifications page of our online catalog. And if you don’t find what you are looking for, just send us a request. We often synthesize special, non-catalog requests. Contact us at noncat@idtdna.com.


 Processing
Processing