Quick facts
Availability: DNA and RNA
Location: 5', internal, and 3'
Scales: 100 nmol to large scale
Purification: Standard desalt or HPLC
Ordering (modification codes): /5dSp/, /idSp/, /3dSp/
DNA abasic sites
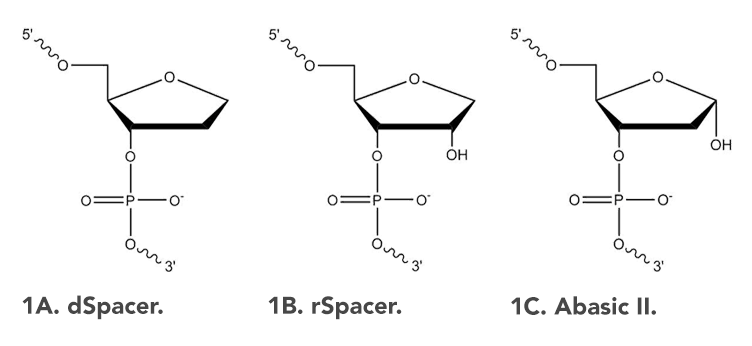
The dSpacer (1,2'-Dideoxyribose) modification (Figure 1A) can be used to insert a single base space into a DNA oligonucleotide sequence. It replicates the loss of any base pairing ability by a nucleotide, which can occur naturally through depurination or other mechanisms [1]. This modification can be useful when examining DNA damage and repair.
dSpacer can also be referred to as an abasic site, tetrahydrofuran (THF), or apurinic/apyrimidinic (AP) site. It may be incorporated into an oligonucleotide sequence internally, or at the 5' or 3' end of a sequence.
To order a dSpacer modified oligonucleotide, go to the Custom DNA Oligos ordering page and enter your sequence using the IDT ordering symbol shown in the blue Quick facts box above to place this abasic site into the desired location within your sequence. You can request dSpacer modified oligonucleotides with standard desalt or HPLC purification.
RNA abasic sites and “unstable” DNA abasic II sites
Additionally, RNA abasic (rSpacer) and the unstable Abasic II modifications are available as non-catalog offerings from IDT. The rSpacer modification (Figure 1B) creates an RNA abasic site with an RNA backbone.
The Abasic II modification contains a hydroxyl group at the 1' position (Figure 1C). The hydroxyl makes this DNA modification more reactive than the dSpacer [2]. We provide oligos with the Abasic II modification in the protected form; therefore, such modified oligos require a deprotection step before use. A deprotection protocol is provided by Glen Research here, on the second page of their published technical bulletin.
To order oligos containing rSpacer (RNA abasic) or abasic II modifications, contact us with your name, organization, and sequence designs. Oligos containing RNA abasic sites, or abasic II sites, can be ordered with HPLC purification. Alternatively, they can be ordered with standard desalt, providing other modifications in the sequence do not require HPLC.