Quick facts about N6-methyladenosine (m6A)
Availability: RNA
Location: 5′, internal, and 3′
Scales: 250 nmol to large scale
Purification: Standard desalt or HPLC
Ordering: /iN6Me-rA/
A popular off-catalog oligo modification
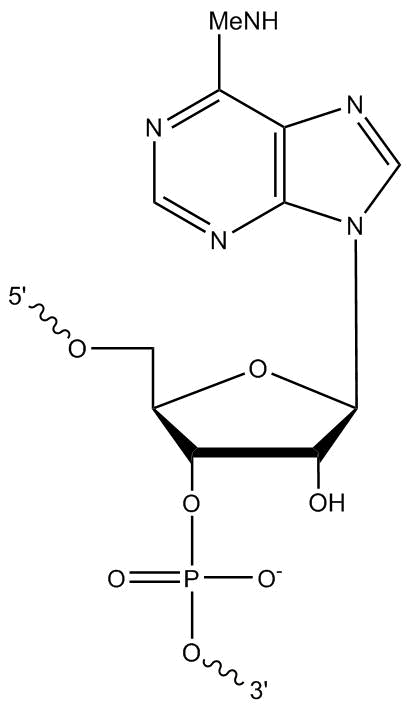
We welcome requests for oligo modifications that do not appear in our website listing. These are regularly reviewed for approval by our specialty oligo synthesis committee, with some made available for quoting and ordering online directly. One popular modification is N6-methyladenosine (m6A). This is a modified RNA base with a methyl group at the N6 position (Figure 1).
First described in the 1970s, m6A is a prevalent internal (non-cap) mRNA modification found in many eukaryotes [1]. It also occurs in other RNA species (tRNA, rRNA, snoRNA, lncRNA) and in a wide variety of organisms, including insects, plants, and yeast [2–5]. It is essential for cell viability and development [4–6], and has been associated with mRNA localization, structure, stability, splicing, and translation [7].
In humans, this methylation event is a natural post-transcriptional modification carried out by a complex consisting of METTL3 and METTL14 methyltransferases and uses the consensus sequence [G/A/U][G>A]m6AC[U>A/C] [2,3]. Human and mouse transcriptome-wide analysis studies and mapping by m6A-CLIP/IP have revealed that the majority of m6A modifications are associated with regions of high evolutionary conservation and are preferentially located in the most 3' exon, suggesting the potential for 3'UTR regulation, including alternative polyadenylation [2,8].
Demethylation of m6A modified bases also occurs naturally within cells. The fat mass and obesity associated (FTO) gene encodes an enzyme that removes the methyl group from this modified base, and overexpression of FTO results in a lower level of m6A in mRNA [2]. The AlkB homolog 5, RNA demethylase (ALKBH5) gene encodes another RNA demethylase responsible for specific demethylation of m6A residues.
How to order m6A modifications
To order RNA oligonucleotides containing m6A modifications, use /iN6Me-rA/ to specify this nucleotide in the sequence you provide. Oligos containing this modification can be ordered with HPLC purification, or with standard desalt, provided that other modifications in the sequence do not require HPLC.
Learn about other oligo modifications to facilitate your research
See what other modifications are available from IDT. You can find a list of standard modifications we make to oligonucleotides on the Modifications page of our online catalog. And if you don’t find what you are looking for, just send us a request. We often synthesize special, non-catalog requests. Contact us to learn more.