Quick facts
Availability: DNA and RNA
Location: 5', internal, and 3'
Scales: 100 nmol to large scale
Purification: Standard desalt or HPLC
Ordering (modification codes): 5Super-dT/, /iSuper-dT/, /3Super-dT/
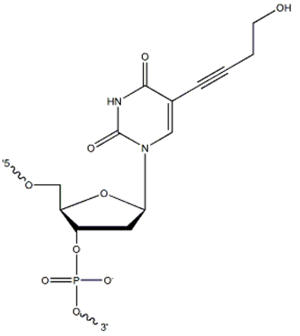
Super T (5-hydroxybutynyl-2'-deoxyuridine; Figure 1) is a modified dT base that can be included in an oligonucleotide primer and probe sequences to stabilize their hybridization [1,2]. Super T base-pairing with dA during primer or probe hybridization provides a ~2°C increase in melting temperature (Tm) per Super T insertion [1]. This Tm increase can be important when working with short primers or probes that are AT-rich.
The Super T modified base is extended normally by polymerases, including Taq polymerase.
How to order
To include Super T bases within your oligo, enter the IDT ordering symbol (/5Super-dT/, /iSuper-dT/, or /3Super-dT/) in the desired location within your sequence. You can request Super T modified oligonucleotides with standard desalt or HPLC purification.