It is frustrating when your eagerly anticipated PCR data show unexpected amplicons. You know your primers are correct, you are certain of the amplicon length, and you definitely used the correct template. So what could be wrong? Most likely, you are facing PCR contamination. There can be various sources of contamination during PCR, leading to myriad observations that may require troubleshooting. A common observation is excessive or unexpected signal, typically caused by contamination of reagents with template, DNA contamination, or amplicon contamination.
Here, we provide some essential tips on how to prevent PCR contamination.
Protect reaction setup from DNA contamination in PCR
Use DNase to degrade genomic DNA before performing reverse transcription. If the aim of your experiment is to measure RNA expression, treat your RNA sample with DNase, and then heat inactivate the DNase before performing reverse transcription.
Design your assays to span exon junctions. To minimize the risk of amplifying contaminating genomic DNA, whenever possible, ensure that your primers span 2 exons (Figure 1).
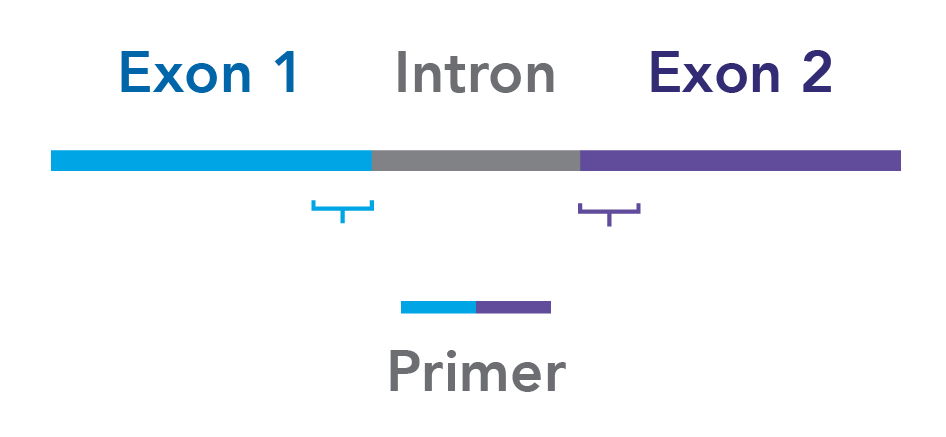
Always incorporate a “no-RT” (–RT) control in your experiment. An –RT control is created by omitting reverse transcriptase in the reverse transcription step and helps to identify genomic DNA contamination in an RNA preparation. Perform this control for any assay that might amplify genomic DNA.
Reduce the risk of cross-contamination from the amplicon/template and previous reactions
Use a unidirectional workflow during experiment setup. Separating pre- and post-amplification areas is key to preventing contamination. Prepare your PCR master mix in a template-free room (see next bullet) using reagents that never come into contact with potential sources of contamination. Maintain a separate area for analyzing PCR amplicons.
Maintain a clean room for preparing master mixes. Prepare your PCR master mix in one room, and then add your template in a separate room to avoid introducing template into the clean room. Prepare components using a benchtop hood when possible to minimize risk of contamination; a benchtop hood is also simple to decontaminate if needed. Keep enzyme mixes, water, primers and probes, pipettes, tubes, tips, and plates in a room where template is not isolated or stored.
Regularly decontaminate equipment. Clean pipettes and other non-porous surfaces with 5% bleach solution to degrade any DNA that may be present. It is easiest to use a spray bottle for this purpose. Leave the solution on the surface for a few minutes to ensure complete degradation of DNA. Alternatively, you can use UV sterilization to decontaminate equipment, including tubes, racks, and pipettes.
Use positive displacement or filter tips for reaction setup. Pipettes are a common source of contamination by aerosols. Filter tips provide a barrier between the pipettes and the liquid being measured, preventing the transfer of aerosols into samples and reagents. Positive displacement pipettes have no air interface between the piston and reagents being measured and, therefore, limit the risk of aerosol contamination.
Exercise general good laboratory practice
Store oligonucleotides in aliquots. We have found that multiple freeze-thaw cycles do not adversely affect the quality of our PCR oligo stock solutions. However, to minimize the risk of contaminating stock solutions, store your oligos at –20°C in single-experiment aliquots. Storing your oligos in aliquots also allows you to repeat the experiment with fresh material if contamination is detected in an experiment using those oligos.
If unexpected amplification in your PCR is confirmed to be due to contamination
Replace all reagents and stock buffers. Discard all existing reagents and repeat the experiment with fresh stocks.
RUO—For research use only. Not for use in diagnostic procedures. Unless otherwise agreed to in writing, IDT does not intend for these products to be used in clinical applications and does not warrant their fitness or suitability for any clinical diagnostic use. Purchaser is solely responsible for all decisions regarding the use of these products and any associated regulatory or legal obligations. RUO23-1864_001

