Frequently asked questions
Our Scientific Applications Support team has assembled a list of frequently asked questions to help you find answers quickly. Filter using one or more categories to focus on specific topics, or use the search bar to perform a text search.
Search all FAQs:
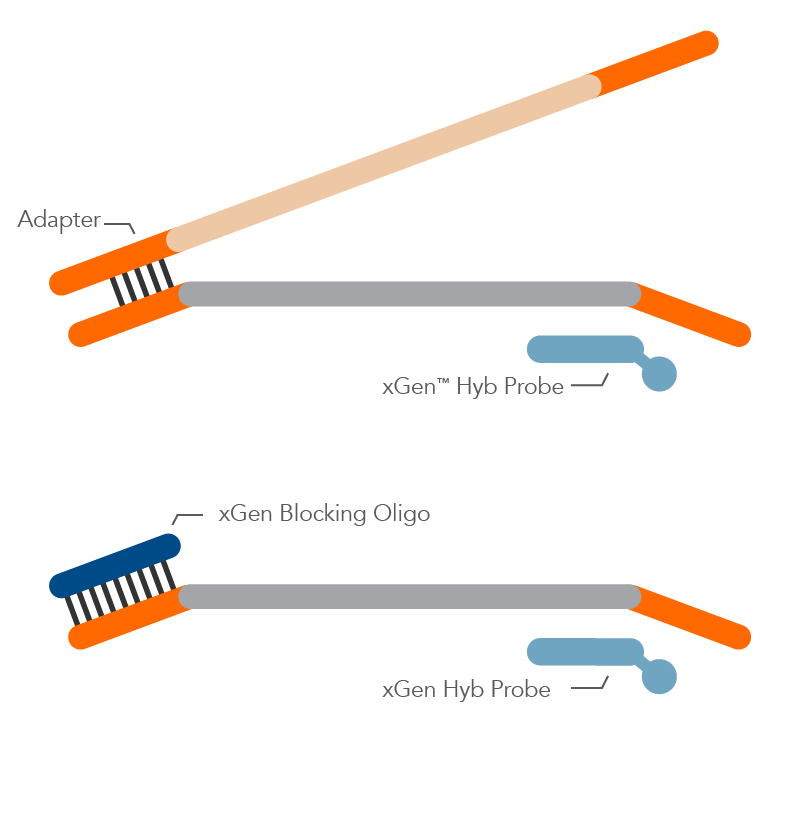
What is the difference between universal and standard blocking oligos (used in NGS experiments)?
Standard blocking oligos are designed for specific barcode sequences. While these designs can be effective in blocking daisy-chaining between fully indexed library molecules, it can be quite cumbersome for the researcher to manage individual blocking oligos that mate with specific indexes, as the researcher expands the number of samples to multiplex together.
The xGen™ Universal Blocking Oligos solve this problem of managing a 1:1 blocker to adapter matching by using a proprietary oligo design to allow our universal blockers to block effectively across ALL index designs of a particular length, while also boosting the on-target percentage rate as compared to standard blocking oligos.


 Processing
Processing