Programming complex oligo structures for new applications
In 1953, when James Watson and Francis Crick described the structure of DNA, they quickly realized its importance for storing genetic information [1]. In the 60 years since Watson and Crick made their discovery, chemists and biologists have found many ways to manipulate DNA—and more recently RNA—that rely on the basic principles of A-T and G-C base pairing and the joining of 5’ phosphate to 3’ hydroxyl. This insight has driven much of modern biological science and medicine.
However, the properties of DNA also make it interesting for other purposes. Dr Peng Yin, an assistant professor of systems biology at Harvard Medical School and a core faculty member of Harvard’s Wyss Institute for Biologically Inspired Engineering, has found new ways to use the information stored in DNA base pairing to instruct DNA to create complex shapes, including 3D structures. This new 3D nano-structure construction method, referred to as “DNA bricks”, opens up entirely new applications for synthetic DNA and RNA.
How DNA bricks work
A DNA brick is a relatively simple, single-stranded, 32-base DNA oligonucleotide. Each brick has 4 consecutive 8-base domains, and they assemble at right angles to complementary interlocking oligos guided by these 8-base domains (Figure 1).
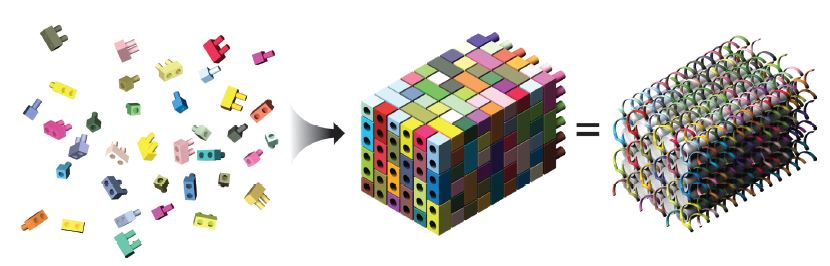
These bricks can then be assembled into a molecular canvas that is represented as a 10 x 10 x 10 cubic “voxel” (volumetric pixel element) structure in the software designed by the Yin lab, with the bricks mapping to specific voxel locations. A 3D shape is created by withholding individual voxels from the brick, and the software then determines which oligonucleotides are required to form the resulting 3D canvas structure (Figure 2) [2].
Dr Yin explains that when they first began making 2D shapes, his team would screen every sequence for energetics, base composition, and similar binding affinity, and then test them for potential cross reactivity. However, the researchers have since discovered that this analysis is not really necessary for the particular structures that they built. The assembly is impressively robust even when choosing the interacting oligo domains at random. Dr Yin says that this robustness is most likely influenced by the fact that each oligo within the brick is positioned in the final assembly by the cooperation of 4 individual DNA elements working to locate it to the proper location.
The lab has written software to automate most of the design process. Combined with the active self assembly of the DNA oligos, DNA bricks are relatively easy to design and use.

Manual pipetting vs. automated liquid handling
As was previously mentioned, a complete molecular canvas structure is composed of 1000 voxels. For each distinct 3D shape formed, the scientists must create a custom pool of the required oligos from their shape oligo library. That amounts to a lot of pipetting. Dr Yin told us that early on in their work, “We used to have people in the lab do the pipetting by hand, which caused a certain amount of complaints.” The researchers now use custom protocols on an automated liquid handler to do this work for them. The scientists simply upload a drawing of the final desired shape and the software is able to choose the appropriate oligonucleotide strands and add them to the required oligo pool for that shape. To the appreciation of everyone in the lab, they were able to automate this process before they started studying the 1000-voxel molecular canvas.
What are the applications for DNA bricks?
The unique physical properties of DNA bricks allow them to be used in ways that are entirely new for DNA, and Dr Yin and his collaborators have already published some interesting applications. For example, in a recent paper, Dr Yin and his collaborators at the Massachusetts Institute of Technology used 2D, single-stranded tiles, similar to the 3D bricks, to mask patterns in grapheme. The DNA was then coated with gold, after which the uncoated graphene was chemically dissolved. This procedure could eventually be used to reliably create small circuits on a nanometer scale for a variety of applications [3]. Similarly, the DNA bricks can serve as a scaffold for growing other molecular matrices, such as those formed from SiO2, which have many material applications [4].
The DNA bricks can also be easily modified with other functional molecules, such as fluorophores. Dr Yin and his group have used DNA origami methods to create multichannel, fluorescent barcodes from DNA rods that are assembled in a slightly different manner from the DNA bricks. By using 3 colors and unique spatial arrangements, Dr Yin and his colleagues were able to create 216 unique visual barcodes that could then be distinguished by fluorescence microscopy [5]
The future of DNA brick technology
This is still a relatively new area of DNA science, and Dr Yin has many ideas for possible DNA brick applications. The research team continues to work on scaling up the size of the structures and their capacity to produce them. Currently the lab is able to make shapes of ~5–10 megadaltons. They would like to continue scaling up so that they can produce structures as large as 100 megadaltons or even 2 gigadaltons!
Additionally, the lab regularly reviews new potential applications; for example, encoding interesting dynamics into the structures. They have already published a paper on engineering the self-assembly pathway of the DNA structures using programmable DNA hairpins that can be designed to interact with each other in a kinetically controlled fashion [6]. These DNA hairpins can be viewed as “dynamic DNA bricks”. Dr Yin also explained, “Rational design of chemical functions will continue to be challenging because while we have control on a nanometer scale, we do not have the ability to make manipulations on the angstrom scale at which enzymes function. So you can’t really rationally design something that functions like a restriction enzyme, at least not yet. However, you can affect nanometer scale interactions by using the DNA structures as scaffolds to bring together 2 other functional molecules such as protein enzymes.”
In addition to catalysts, DNA bricks have the potential to serve as scaffolds for other functional molecules to create “biofactories” that can produce other useful molecules.
Can we use other biopolymers?
When asked about protein, Dr Yin stresses that “biology relies on digital molecular systems and proteins are indeed digital molecules. However, while proteins are quite fascinating, our ability to rationally analyze and design proteins is currently still too limited to be able to program them in the same manner as we can now program nucleic acids.”
Therefore, says Dr Yin, “Because of their very simple base pairing rules we can easily encode complex geometry and dynamic behavior in DNA and RNA molecules. DNA and RNA are quite unique in this way. However, the general principle (of digital information–directed molecular behavior) that we demonstrated should be generalizable to other digital molecules. For example, it would be really fascinating to design DNA-like synthetic polymers or peptides that can follow the digital base pairing rules and use them to engineer complex shapes or dynamics.”
Origami method vs. bricks
It is easy to confuse the DNA brick method with DNA origami, which has also recently been published and described in detail in the news. However, there are some key differences.
DNA origami uses a long, single strand of DNA as a scaffold strand. Functionality and shapes are created by adding short DNA “staple” fragments. In addition, you can use the properties of the staple strands to fold the scaffold strand into shapes. The choice of scaffold strands has been limited because, until recently, it was difficult to obtain long strands of synthetic DNA. Viral genomes have been used as scaffolds, but their sequence is not controlled by the researcher. The final structure is made up of staple strands attached to a core scaffold strand, and until the DNA bricks method, it was the only way to make complex megadalton scale structures [7].
Bricks, on the other hand, are highly modular and much easier to implement. Individual oligos can easily be included, excluded, or replaced. Because there is no long scaffold strand, the bricks method is much more flexible. Once you have a complete 3D brick design, you can create new shapes by simply treating each oligo as an individual voxel and virtually carving the shape by removing specific voxels. In addition, because the bricks are completely synthetic, you can affect their other properties. For example, instead of using D-DNA, you can use L-DNA, which is resistant to enzyme degradation and has lower immunogenicity. L-DNA structures may therefore be useful for drug applications. And, of course, these same DNA bricks methods can be applied to make RNA structures.
IDT oligos inside
Clearly, the DNA bricks projects use a lot of oligonucleotides, both with and without modifications. Dr Lin says that he has personally been using IDT oligos since his graduate work, more than 10 years ago. He has been very happy with the quality of IDT oligos, and notes that when they once tried another supplier, those oligos did not produce structures as well as the ones from IDT.


 Processing
Processing


