xGen Lockdown Probe Pools
Custom target enrichment panels for maximum flexibility and performance
xGen Lockdown Probe Pools contain individually synthesized Lockdown Probes pre-formulated to deliver consistent, convenient, best-in-class performance.
- Achieve higher performance with individually synthesized and quality-controlled capture probes
- Develop cost-effective custom panels that are delivered quickly (7–10 business days for orders <2000 probes)
- Optimize, expand, or combine custom panels
Ordering
xGen Lockdown Probe Pools
Use our Design tool to select predesigned or create custom xGen Lockdown Probe Pools. If you already have target capture probe sequences, select Order my designs to upload your sequences.
16 reactions
96 reactions
4 x 96 reactions
For legacy xGen Lockdown Probe customers and customers who require large quantities of delivered probes, please contact CustomQuotes@idtdna.com.
Product details
xGen Lockdown Probes are individually synthesized 5′-biotinylated oligos for hybridization capture enrichment in next generation sequencing. xGen Lockdown Probes are produced using the same industry-leading, high-fidelity synthesis chemistry developed for Ultramer Oligonucleotides.
These probes can be used for creating custom capture panels that can be optimized, expanded, and combined with other panels. xGen Lockdown Probe Pools can also be used to enhance the performance of existing capture panels, rescuing poorly represented regions, such as areas of high GC content.
With xGen Lockdown Probes, you can:
- Design probes for any sequence in any species using your specified parameters for target definition, probe length, and tiling density
- Add probes for custom targets like untranslated regions (UTRs), introns, and fusions to complement human xGen Predesigned Gene Capture Pools
- Easily transition to high volume applications with custom production scales
Use gene symbols, gene IDs, and BED files to identify human, mouse, and rat gene targets. Alternatively, the design tool also accepts RefSeq accession numbers or FASTA-formatted sequences.
Performance
High target coverage
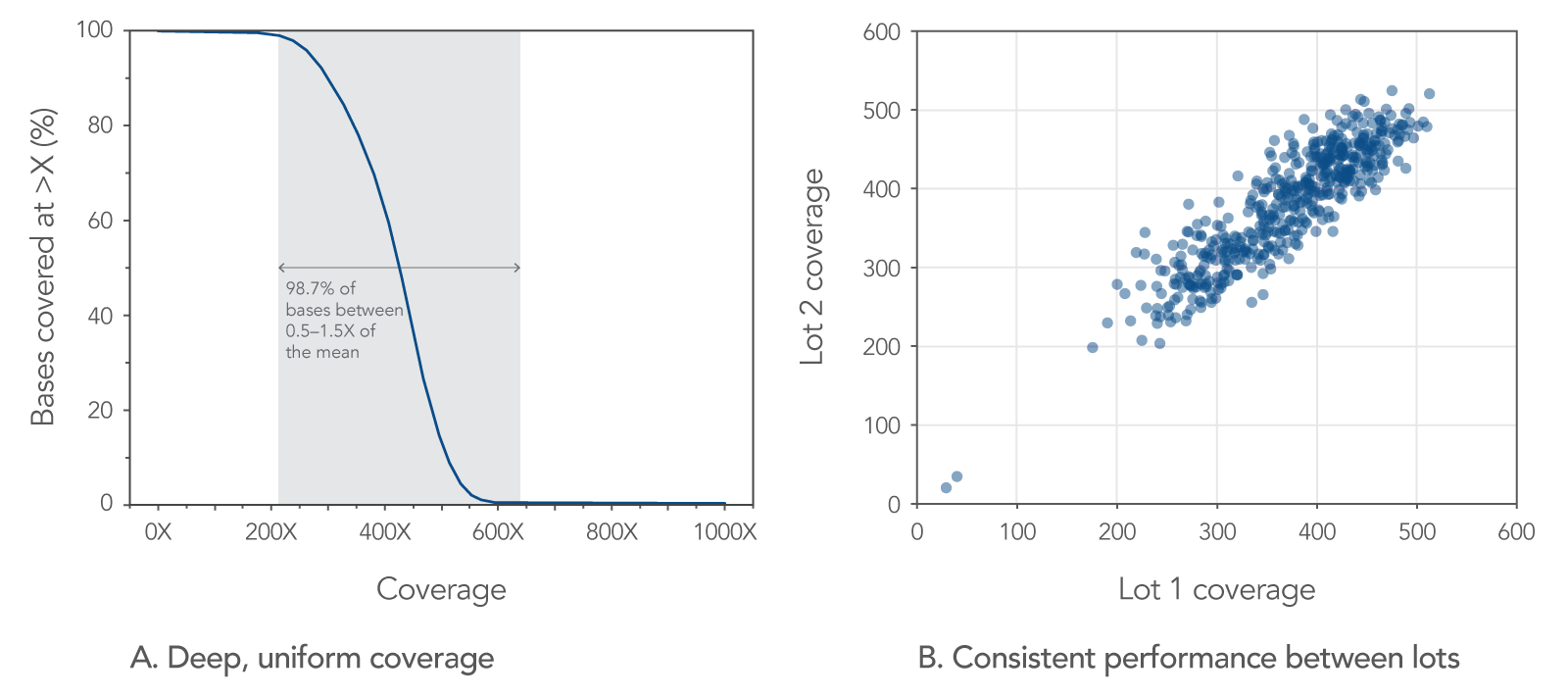
Capture panels created with xGen Lockdown Probe Pools provide deep, uniform coverage of the targeted regions (Figure 1), because all probes are present at equimolar concentrations. Individual synthesis of Lockdown Probes also delivers reliable batch-to-batch performance.
Figure 1. Consistently deep, uniform sequence coverage with xGen Lockdown Probe Pools. A DNA library created from human genomic DNA (Coriell) was enriched for an 88 kb target region, using an xGen Lockdown Probe Pool. The enriched libraries were sequenced on an Illumina NextSeq® instrument and subsampled to 600K reads. The data show (A) deep, uniform coverage and (B) consistent mean target coverage for two synthesis lots of 1000 probes. The mean of duplicates for each synthesis lot are plotted for the scatter plot. Coverage and target depth were calculated with Picard and BEDTools, respectively.
Efficient capture across a range of GC content
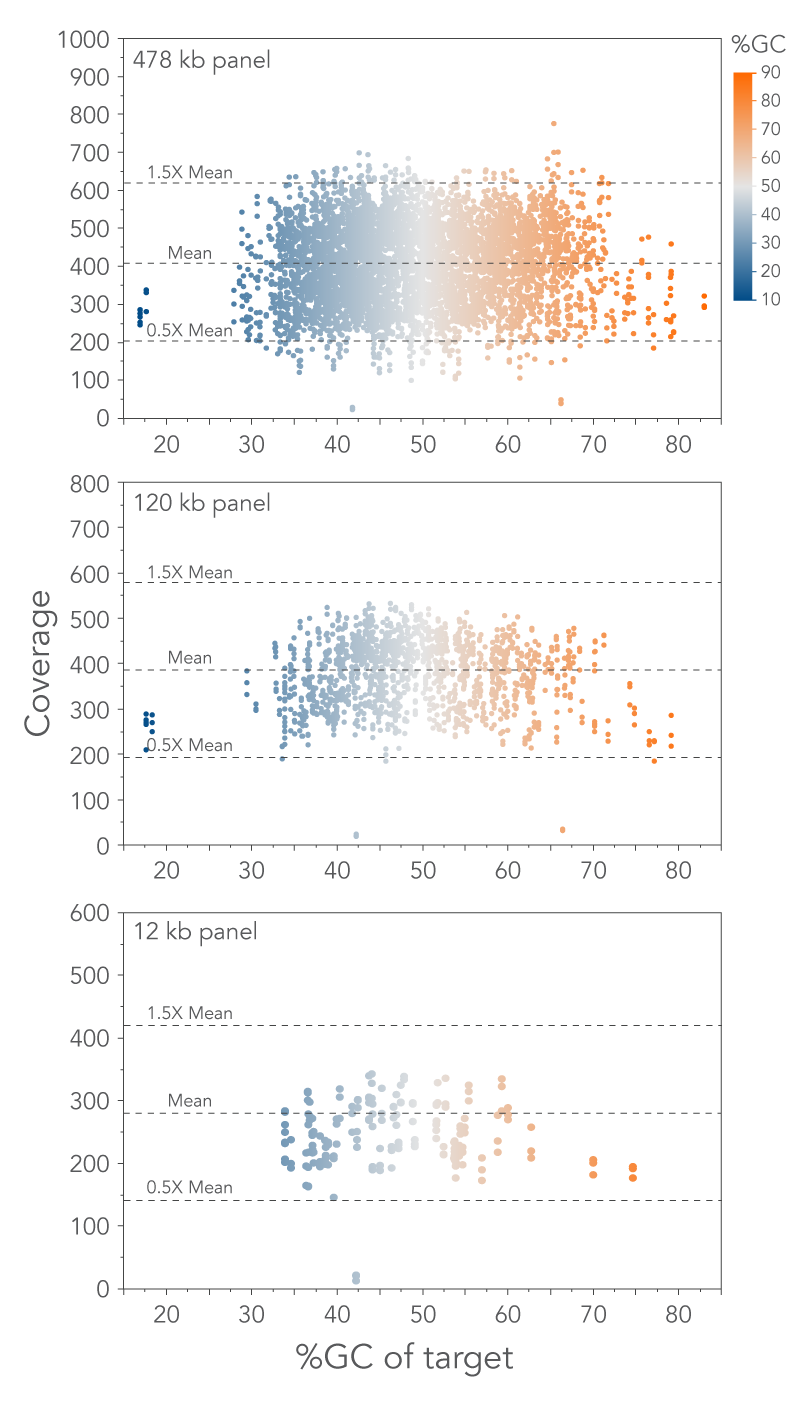
xGen Lockdown Probes have low bias across targets containing a wide range of percent GC.
Figure 2. Minimize GC bias using xGen Lockdown Probe Pools. Targets are uniformly covered across a wide range of GC content. Libraries were prepared from Coriell NA12878 genomic DNA and captured with 3 Custom Lockdown Probe Panels. Target coverage verses target GC percentage is shown for each panel: (top) 369 kb target space with a 478 kb probe footprint, (middle) 90 kb target space with a 120 kb probe footprint, and (bottom) 9 kb target space with a 12 kb probe footprint.
Customize your panel by adding lockdown probe pools
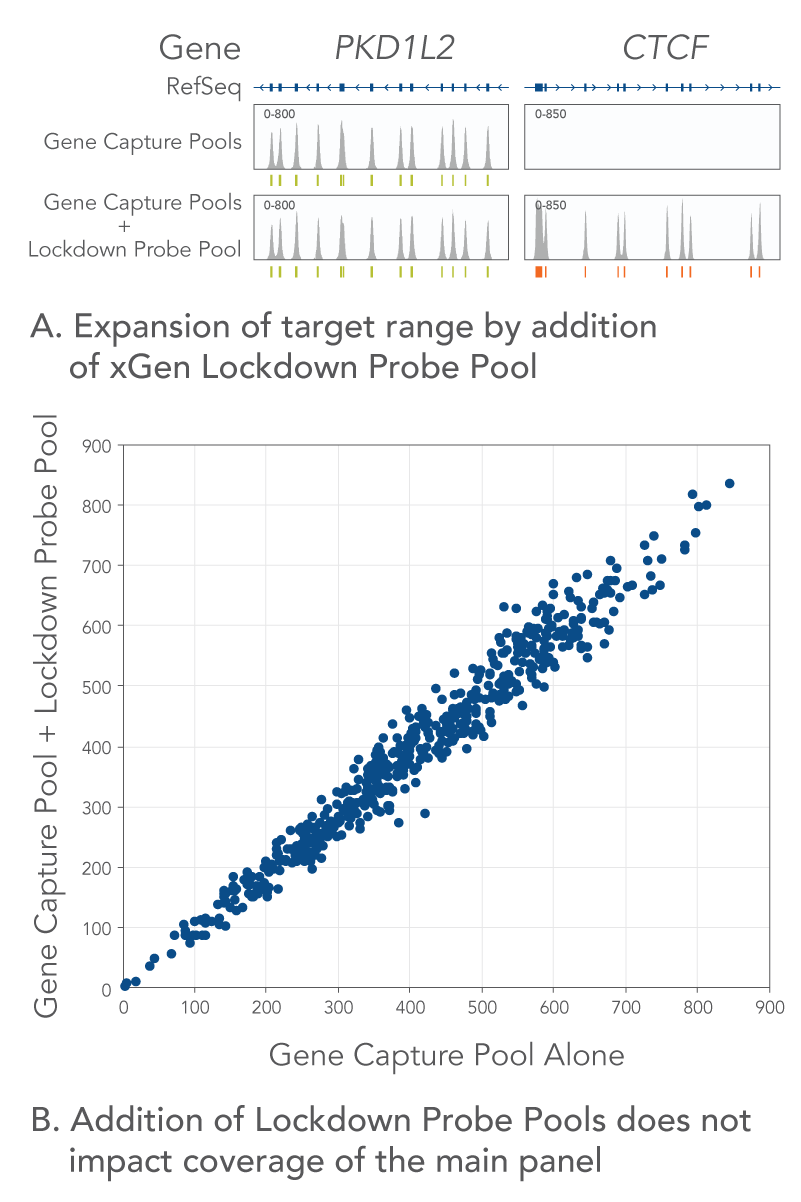
xGen Lockdown Probe Pools are formulated at known equimolar concentrations, so they are modular. Probe pools can be added to an existing panel to expand the target region without impacting coverage of the original panel.
Figure 3. Add custom targets to an existing panel. xGen Lockdown probes can be added to target regions of interest like UTRs, introns or fusions to customize an existing panel. Libraries were prepared from Coriell NA12878 genomic DNA and captured using either a Gene Capture Pool (1232 probes targeting 27 genes covering a 115 kb region) or an xGen Lockdown Probe Pool targeting 14 additional genes (9 kb) was spiked into the Gene Capture Pool for capture. Probe regions are indicated. (A) Representative IGV screenshots are shown. (B) Coverage of xGen Predesigned Gene Capture Pool targets, with and without the spike-in of Lockdown Probe Pools, demonstrates that adding additional Lockdown probes does not impact coverage of the main panel.
Improve coverage of array-synthesized baits
xGen Lockdown Probes can be added to array-derived baits to enhance the coverage of GC-rich targets, such as first exons (Figure 4).
Figure 4. Spiking in individually synthesized oligonucleotide probes improves sequencing coverage of array-derived RNA baits. Individually synthesized 5′-biotinylated xGen Lockdown Probes were spiked into commercial, array-synthesized RNA baits to improve coverage of areas with high GC content. Either (A) 1000 xGen Lockdown Probes targeting a ~133 kb region, or (B) 3 xGen Lockdown Probes targeting a single exon, were mixed with 1.1 Mb RNA baits and hybridized for 24 hr, followed by target capture using streptavidin. Enriched targets were sequenced on a HiSeq® 2000 system (Illumina) using 49 x 49 paired-end reads. Lockdown probes improved general coverage of (A) GC-rich targets and (B) single GC-rich exons.
Resources
User guides and protocols
Legacy protocols
Legacy resuspension calculator
Calculate resuspension volumes and number of reactions based on ordering scale and number of probes


 Processing
Processing



Leave a comment