xGen Hybridization and Wash Kit
Optimized reagents for the xGen hybridization capture workflow
The xGen Hybridization and Wash Kit comprises buffers, Cot DNA, and streptavidin beads required for hybridization capture of DNA. The kit has been developed to work with xGen Lockdown Probes and Panels and xGen Blocking Oligos for a complete, high quality target enrichment solution. This workflow is compatible with NGS libraries prepared using ligation-based techniques, such as TruSeq® library kits, and Nextera® DNA library preparation kits.
- Achieve uniform coverage and robust capture performance across a broad range of xGen hybridization probe panels
- Generate highly reproducible targeted NGS data with quick and easy workflow results
- Updated, automation-friendly protocol enables various levels of throughput
Ordering
Note: the xGen Hybridization and Wash Kit is required for the updated protocol, xGen hybridization capture of DNA libraries for NGS target enrichment, v1.
For the previous xGen Lockdown Reagents kit, please click here.
Product details
The xGen Hybridization and Wash Kit comprises hybridization and wash buffers, Cot DNA, and streptavidin beads that have been optimized for in-solution target enrichments using xGen Lockdown Probes and Panels. The reagents have not been tested with target capture probes from other manufacturers.
| Description | xGen Hybridization and Wash Kit | xGen Lockdown Reagents* (legacy kit) |
|---|---|---|
| 2X Hybridization Buffer | ✅ | ✅ |
| Hybridization Buffer Enhancer | ✅ | ✅ |
| 2X Bead Wash Buffer | ✅ | ✅ |
| 10X Wash Buffer I | ✅ | ✅ |
| 10X Wash Buffer II | ✅ | ✅ |
| 10X Wash Buffer III | ✅ | ✅ |
| 10X Stringent Wash Buffer | ✅ | ✅ |
| Dynabeads™ M-270 Streptavidin | ✅ | |
| Human Cot DNA | ✅ | |
| Compatible with manual plate protocol | ✅ | |
| Compatible with automation scripts | ✅ |
* The xGen Lockdown Reagents kit is not recommended for use with the updated protocol, xGen hybridization capture of DNA libraries for NGS target enrichment, v1 because it does not contain sufficient volumes for some buffers required for the updated protocol.
xGen Hybridization and Wash Kits are shipped in two boxes, labeled Box 1 and Box 2, which must be stored at different temperatures. Box 1 contains all components except for streptavidin beads and requires storage at –20°C. Box 2 contains Dynabeads M-270 Streptavidin and should be stored at 4°C.
For more information on Human Cot DNA and to order additional quantities, click here.
Library amplification primers for use with Illumina adapter P5/P7 libraries are available here.
Technical overview
The xGen Hybridization and Wash Kit has been developed and optimized for use with xGen Lockdown Probes and Panels and xGen Blocking Oligos. The kit is compatible with the protocol, xGen hybridization capture of DNA libraries for NGS target enrichment, v1, to provide a complete, high-quality hybridization capture solution for targeted NGS.
The xGen Hybridization and Wash Kit and the protocol, xGen hybridization capture of DNA libraries for NGS target enrichment v1, replace the xGen Lockdown Reagent kit and the protocol, Hybridization capture of DNA libraries using xGen Lockdown Probes and Reagents, v3.
Automation
We have partnered with several instrument vendors to develop automation scripts for the xGen capture protocol on liquid handling robots. Scripts are currently available for the following:
- Beckman Coulter Biomek® i5 and i7 Automated Workstations
- Hamilton NGS STAR™ for Library Prep workstation
- Perkin Elmer Sciclone® workstation
Please contact the instrument vendors for more information regarding script availability and installation or contact IDT Customer Care for an updated list of qualified automation scripts for liquid handling robots.
Performance
The xGen Hybridization and Wash Kit and capture protocol has been optimized to work with a broad range of sizes of xGen Lockdown Panels, ranging from small panels (20 kb) to the large xGen Exome Research Panel (51 Mb). On-target rate and coverage uniformity using the xGen Hybridization and Wash Kit for a given xGen panel are reproducible across a variety of target capture scenarios to support various throughput needs (Figures 1–3). The performance data presented were generated on a NextSeq® 500 Sequencing System (Illumina), aligned with BWA (ref) to human reference hg19, and coverage statistics compiled with BEDTools (ref).
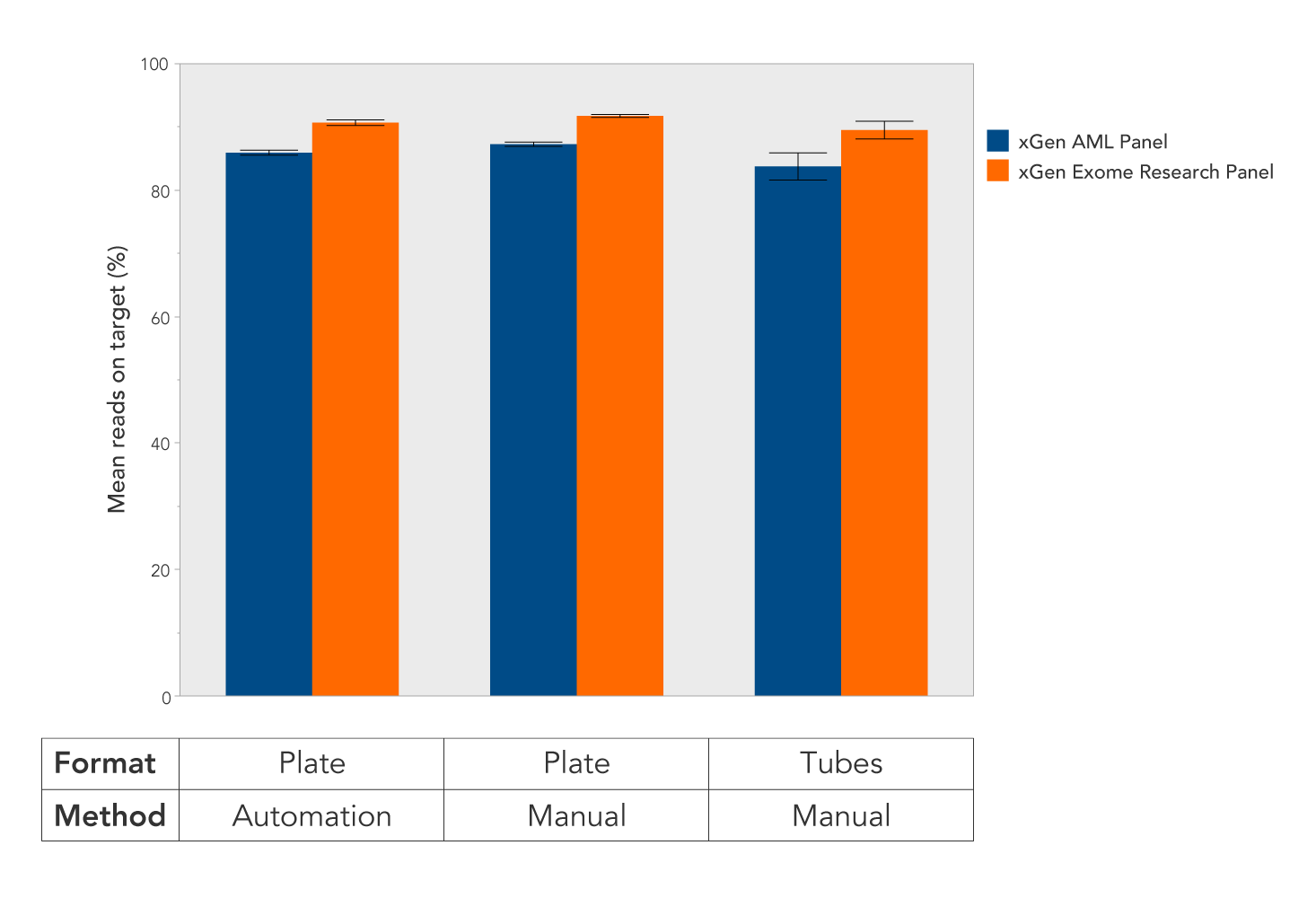
Figure 1. Robust, reproducible on-target rates with different size panels independent of capture format. Sequencing libraries with an average insert size of ~300 bp were prepared from replicate samples of NA12878 genomic DNA (Coriell). The libraries were enriched using the xGen Exome Research Panel and xGen Acute Myeloid Leukemia Cancer (AML) Panel and the indicated combinations of methods (manual or automated) and formats (plate or tubes) (n = 8 samples for each data set). The xGen Hybridization and Wash Kit was used according to the protocol in xGen hybridization capture of DNA libraries for NGS target enrichment, v1. Automated runs were performed using the Perkin Elmer Sciclone robot with automation scripts developed jointly by IDT and Perkin Elmer. On-target rates were calculated using 150-base flanks added to the ends of target regions.
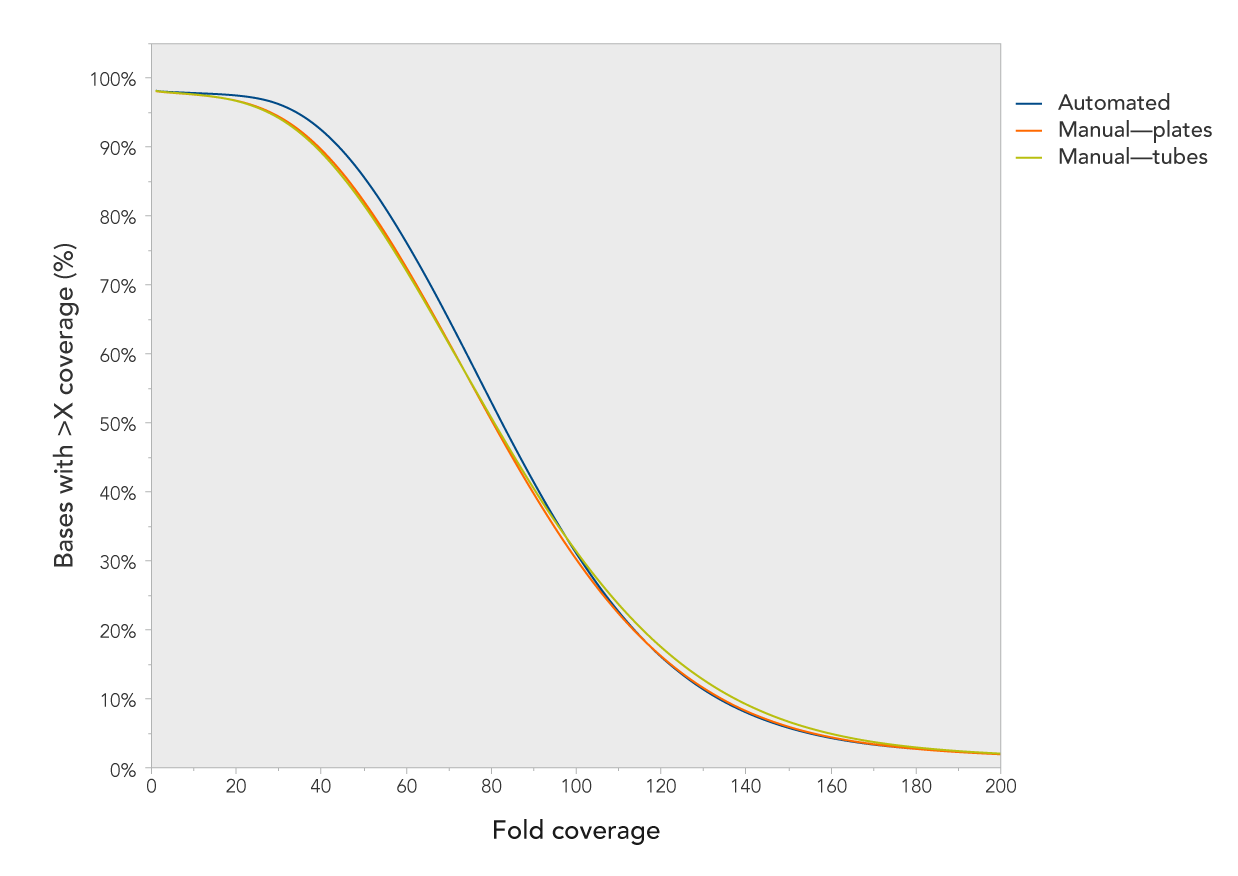
Figure 2. Consistent, uniform coverage using different capture formats and methods. Libraries prepared from genomic DNA NA12878 (Coriell) were enriched using the xGen Exome Research Panel by either manual (tubes and 96-well plate) or automated (96-well plate) methods. More than 94% of bases were covered at 30X for both methods, using 25 million paired-end reads per sample sequenced 2 x 150 bp.
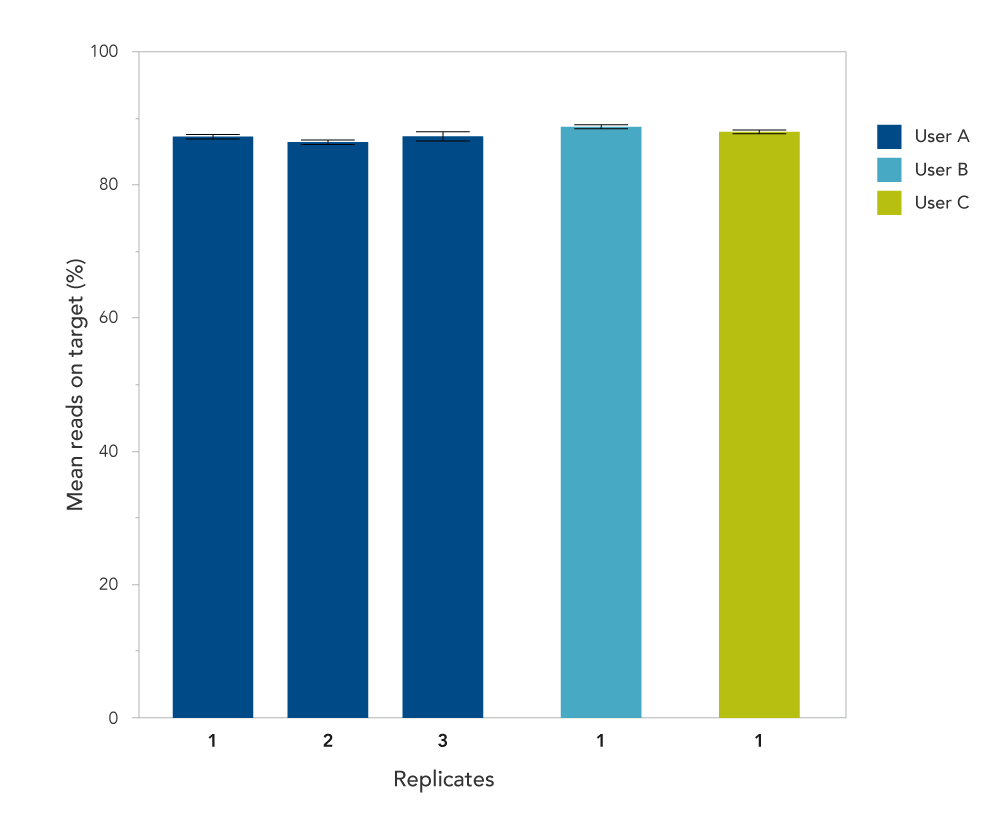
Figure 3. Reproducible on-target rates from enrichment in 96-well plates. DNA libraries prepared from NA12878 (Coriell) were enriched using the xGen AML Cancer Panel 1.0 with the xGen Hybridization and Wash Kit. Target capture was performed by multiple users (A–C), using the manual protocol for 96-well plates. User A also performed independent replicates on different days. On-target values (with 150 bp flank) were averaged for each data set (n = 8).


 Processing
Processing