OligoAnalyzer™: Primer analysis tool
Understand the expected properties of your oligos before you order them.
- Calculator for GC content, melting temperature (Tm), molecular weight, extinction coefficient, µg/OD, nmol/OD, and more
- Identify secondary structure potential
- Minimize dimerization
- Use NCBI BLAST™

Research-friendly oligo calculator
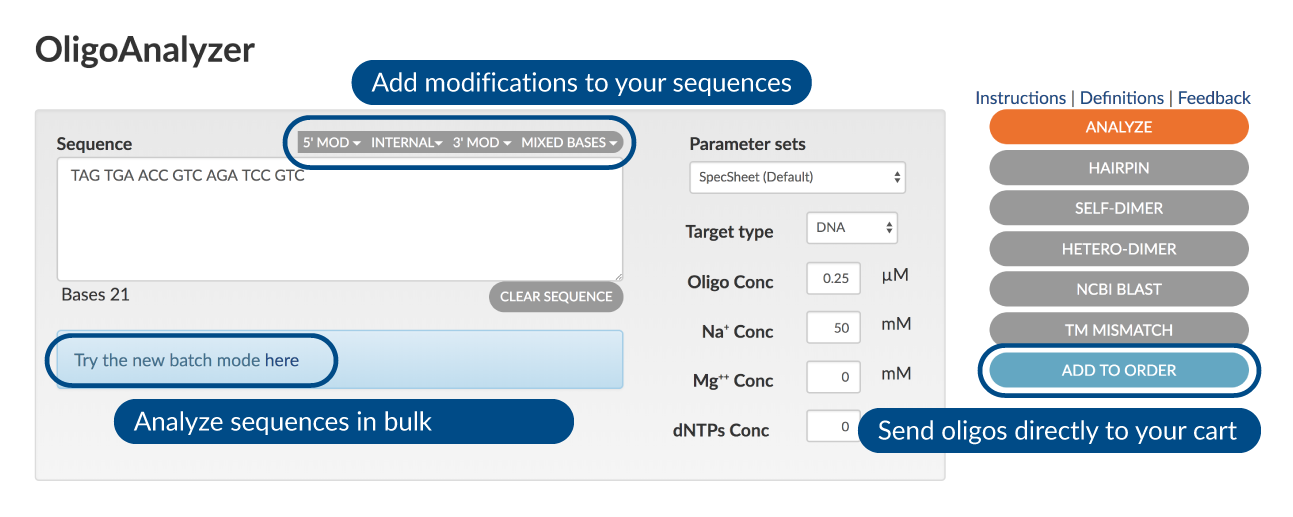
Flexible input and advanced parameters to optimize your custom order:
- Enter your primer or other oligo sequence
- Adjust calculation options if desired
Choose a function:
- Select ANALYZE for easy, one-click access to a Tm calculator, GC content calculator, extinction coefficient calculator, and more.
- Predict oligo secondary structure or possible duplexes with HAIRPIN, SELF-DIMER, and HETERO-DIMER options.
- Search for matches to your input oligo sequence in the NCBI BLAST database.
- Explore mismatch and melting temperature variations with the TM MISMATCH option.
New to the OligoAnalyzer tool?
Our video will introduce you to the basics of our oligo analysis tool and get you up and running quickly. We’ll go over the various functionalities available in the tool, using example sequences.
Creating optimized custom oligos just got easier
Enter your sequence
- DNA
- RNA
- Locked nucleic acid bases
- Modifications
- Mixed bases
- Phosphorothioated bases
Adjust calculation options
- Hybridization target type
- Oligo concentration
- Na+ concentration
- Mg2+ concentration
- dNTP concentration
- Use SpecSheet or qPCR preset values
Choose a
function
- Standard analysis
- Hairpin
- Self-dimer
- Hetero-dimer
- NCBI BLAST
- Tm mismatch
Resources
Web resources
OligoAnalyzer user manual
In-depth information on the tool’s functionality and features.
Help with terminology
A glossary of terms you will see when using the tool.
Need help or inspiration?
If you need further assistance please call, email, or web chat us today.
RUO23-1671_001
