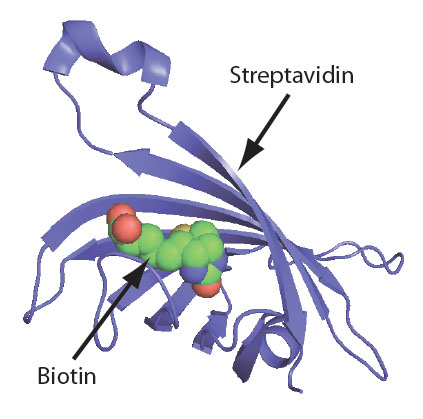
Biotin is an important molecule in molecular biology applications, partly due to its very high affinity for streptavidin and avidin. It is used in mobility shift assays, and for enrichment, purification, and attachment to solid surfaces. Biotin can also be used for tagging target molecules with dye- or enzyme-labeled streptavidin. The affinity of the biotin–streptavidin interaction is extremely strong, with a disassociation constant (Kd) of 10−15 M. Biotin fits inside a ‘pocket’ (Figure 1) of the streptavidin protein [1], creating an enveloping effect which supplements the binding interaction. It is this binding strength that makes biotin a very useful tool for molecular and biomedical research.
Choosing a biotin modification
Biotin can be added to oligonucleotides on either terminus (“standard” Biotin), as well as internally through a modified thymidine residue (Biotin-dT). There are several types of biotin modifications—each with their own benefits—all containing the same functional biotin group.
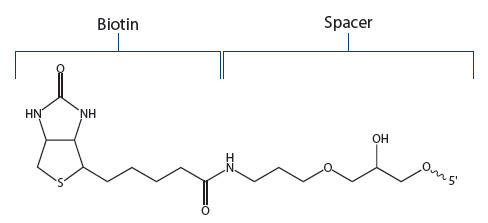
Standard Biotin. IDT offers a biotin modification that is attached to the 5’- or 3’-ends of an oligo using a C6 (standard) spacer (Figure 2). Referred to by IDT as “Biotin”, this version is recommended for most applications.
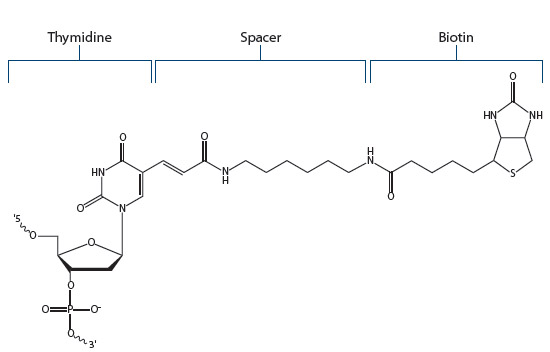
Biotin dT. Biotin-modified thymidine residues (Figure 3) allow the addition of biotin internally within an oligonucleotide. Biotin dT residues can also be added to either end of an oligo.
Biotin-TEG. Biotin-TEG increases the oligo–biotin distance to 15 atoms using a triethyleneglycol (TEG) spacer. Biotin-TEG is commonly used to avoid hindrance issues and can be beneficial for attaching oligonucleotides to nanospheres or magnetic beads.
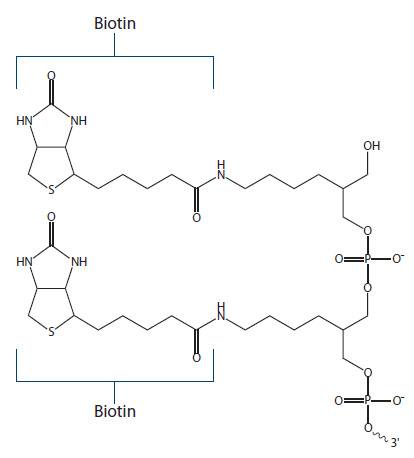
Dual Biotin. Dual Biotin is a modification resulting in two functional biotin groups (Figure 4), which act to increase biotin–streptavidin binding affinity, and are used for applications like Serial Analysis of Gene Expression (SAGE) assays.
PC Biotin and DesthioBiotin-TEG. One of the most challenging, and often frustrating, aspects of applications employing biotin is the nearly irreversible biotin–streptavidin interaction. The bond is stable over a broad pH and temperature spectrum. Conditions necessary to release biotin can be potentially harmful or negatively affect downstream procedures. There are two biotin modifications that provide binding and controlled release:
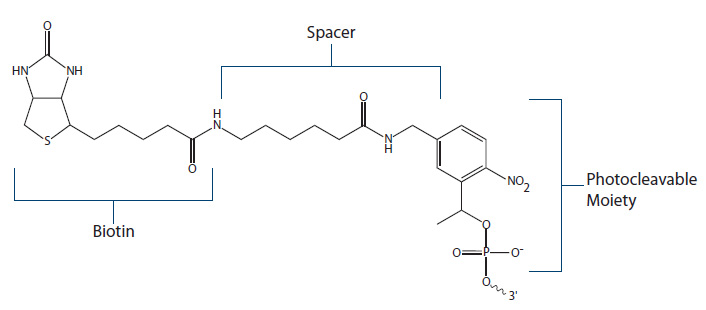
PC Biotin (Figure 5) employs a photocleavable spacer arm which can be cleaved when exposed to UV light of specific wavelength (300–350 nm). One benefit of the PC modification is that upon cleavage, the resulting DNA oligo will have a free phosphate group available for subsequent ligase reactions.
DesthioBiotin-TEG, a biotin analog missing the sulfur atom, is another option for post-binding release. This analog binds tightly to streptavidin, but more weakly than standard biotin. Because of this, rinsing streptavidin bound oligos with buffered solutions containing free biotin will result in the displacement of desthiobiotin with the free biotin, allowing the oligo to be removed and collected [3].
Biotin Azide. Finally, biotin can be added to oligonucleotides either at the 5’ end or internally, using a reactive azide group in a click chemistry reaction (see the article, Oligo modification—post-synthesis conjugation explained, for more information about this reaction).
Ordering modifications from IDT
Biotin modifications are available for ordering from IDT and can be found listed under the 5’, 3’, and Internal Modifications tabs on the oligonucleotide ordering web page. Note that most biotin modifications require a minimum processing scale of 100 nmoles and HPLC purification.
The IDT Oligo Modifications web page provides more structures and information about these modifications for biotinylation. For any questions regarding biotin options from IDT, please Contact us.