Background
Current targeted sequencing strategies may be expensive, time-intensive, or offer low yield or limited read length. There is a need for fast, inexpensive, flexible, but comprehensive, sequencing options.
Nanopore sensing involves embedding a tiny hole, or nanopore, into an electrically resistant, polymer membrane, and using the nanopore to identify molecules that contact it. When a molecule passes through or blocks the nanopore, the current is disrupted. That disruption can be measured. DNA bases, RNA bases, modified bases, proteins, and small molecules can all be identified in this way. A strand of DNA can be sequenced in real time as it passes through the nanopore, allowing for sequencing of much longer reads than possible with Illumina® sequencers.
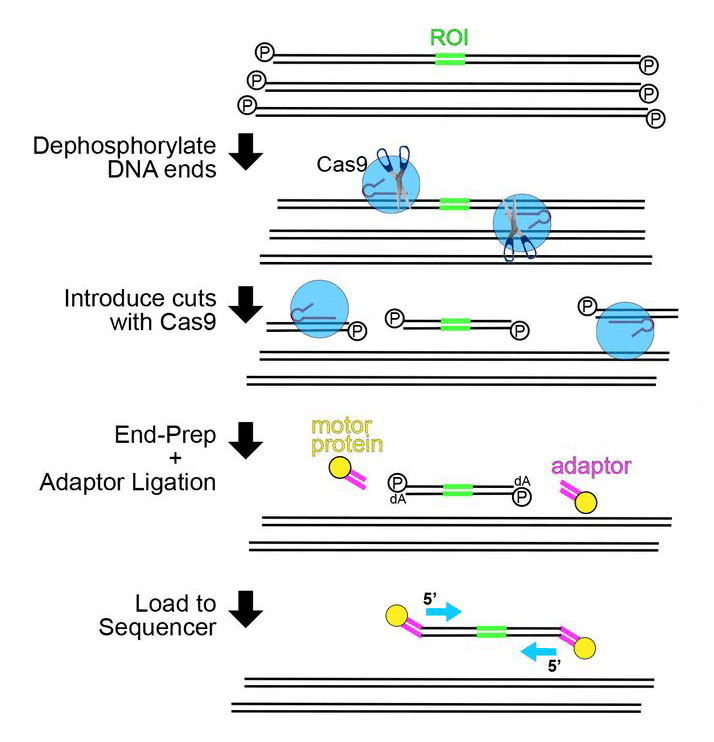
Nanopore Cas9-Targeted Sequencing, or “nCATS,” combines nanopore sequencing technology with Cas9–guide RNA technology for targeted sequencing (Figure 1). Cas9, or CRISPR associated protein 9, is an enzyme that cuts DNA. The Cas9–guide RNA complex, the ribonucleoprotein (RNP) complex, introduces cuts in genomic DNA at specific sites, allowing for sequencing of select regions to reveal DNA methylation, single nucleotide mutations, and structural variations. This method is both scalable and customizable. The whole process requires ~3 µg of genomic DNA and can be completed in a matter of hours, reducing time and cost, and increasing efficiency.
Experiment
Gilpatrick, et al. tested nCATS using genomic DNA (gDNA) from 4 cell lines: the well-characterized GM12878 lymphoblast cell line and 3 breast cell lines (MCF-10A, MCF-7, and MDA-MB-231) [2]. Ten genomic regions, chosen based on existing expression data from these cell lines, were targeted. A custom panel of guide RNAs, selected for optimal on- and off-target editing, was designed using the custom Alt-R™ CRISPR-Cas9 crRNA design tool. RNP complexes were constructed by combining the guide RNA, composed of custom Alt-R CRISPR-Cas9 crRNA and tracrRNA, with Alt-R HiFi Cas9 Nuclease V3. This high-fidelity Cas9 provides highly efficient genome editing with reduced off-target effects. After incubating the RNP complexes with gDNA for Cas9 cleavage, the sequencing adapters were ligated to the resulting fragments, and libraries were prepared. Sequencing was run using the GridION® sequencer (Oxford Nanopore Technologies) (Figure 1). Analyses were performed by both samtools [3] and nanopolish [4].
In subsequent analyses published in Nature Biotechnology [1], Gilpatrick, et al. added to this research by using a multi-gRNA panel and gDNA from a breast cancer cell line xenograft and primary patient tissue. In addition to sequencing on a MinION® device (Oxford Nanopore Technologies), they sequenced on a Flongle® flow cell (Oxford Nanopore Technologies) for comparison. A Flongle flow cell is a smaller, single-use flow cell that adapts to MinION devices for direct, real-time DNA sequencing. WhatsHap, a haplotype assembly tool, assigned reads to parental haplotypes based on single nucleotide polymorphisms (SNPs) revealed by the long-read data [5]. They also performed analyses using Clair and Medaka variant calling tools, which use neural networking algorithms, to compare to samtools and nanopolish. Reads were subsampled to coverages of 300X, 200X, 100X, 50X, and 25X to evaluate the association between variant calling accuracy and coverage depth.
Results
Since the Cas9 RNP directs the sequencing adapter ligation, and the nanopores can sense native DNA strands, PCR amplification is not needed, allowing for more accurate sequencing. Ten- to 300-fold enrichment was achieved in all 10 evaluated regions, resulting in 20X to 800X coverage. Current sequencing technologies are limited by read length; the nanopore sensing allows for longer reads, but that might be at the cost of uniform quantification. Longer strands are associated with less uniform quantification. This is due to the variation in DNA fragment length influencing the concentration of free DNA ends, which impacts the efficiency of adapter ligation.
Nanopore methylation calls were compared to published whole genome bisulfite sequencing data and RNA sequencing data. The nanopore methylation patterns were very similar to the published data. Additionally, methylation signature noise around transcriptional start sites was reduced in the nanopore data compared to the published data. Noise indicates variation. The cleaner signature suggests that regulatory elements, like CPGs, may have less methylation variation, demonstrating an inverse correlation between gene activity and promoter methylation. CPG methylation may be playing a regulatory role in breast cancer.
The experimental strategy revealed differential methylation on a keratin family member gene KRT19, a gene upregulated in breast cancer. KRT29 had allele-specific hypomethylation in the primary patient tumor sample, a feature that would be difficult to evaluate without high-coverage, long-read data produced by this methodology. WhatsHap was able to determine that the hypomethylation occurred on the haplotype with increased copy number.
Structural variation revealed by nCATS was compared to data from the Genome In A Bottle Consortium project from 10x Genomics. The variant caller, Sniffles [6], used on both 10x Genomics and nCATS data, initially failed to recognize reads, identifying them as homozygous. When Gilpatrick, et al., adjusted the acuity of Sniffles, structural variants were accurately called as heterozygous. Using the nCATS method, structural variant calling can be combined with methylation calling to study methylation at deletion points.
The results from single nucleotide variant detection were analyzed by both samtools and nanopolish and compared. Samtools and nanopolish both evaluate variant calling, but nanopolish also takes into account electrical data from the nanopore. While some variants were called with higher confidence than others, overall, nanopolish had higher accuracy and its use resulted in a lower false positive rate than samtools. nCATS, combined with nanopolish, can be used to identify both known and de novo variants.
Additional comparisons presented in Nature Biotechnology demonstrated that the variant calling tool, Clair, had the greatest accuracy at a coverage of 25X and 50X but was not functional over 100X coverage. Medaka’s highest discernment (0.93) peaked at 50X and 100X coverage. Samtools and nanopolish both had their highest accuracies (0.97 and 0.98, respectively) at 200X coverage, indicating that coverage depth should be considered when choosing a variant caller for a particular experiment.
The results presented here demonstrate the viability of nCATS as a reliable sequencing protocol that can be used to reveal methylation, structural variation, and single nucleotide variation. This targeted approach provides a faster, cost-effective method for evaluating genomic or epigenomic variation.
References
- Gilpatrick T, Lee I, Graham JE, et al. Targeted nanopore sequencing with Cas9-guided adapter ligation. Nat Biotechnol. 2020;38(4):433-438.
- Gilpatrick T, Lee I, et al. Targeted nanopore sequencing with Cas9 for studies of methylation, structural variants, and mutations. bioRxiv. 2019; 604173.
- Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27(21):2987-2993.
- Simpson JT, Workman RE, Zuzarte PC, et al. Detecting DNA cytosine methylation using nanopore sequencing. Nat Methods. 2017;14(4):407-410.
- Martin M, Patterson M, et al. WhatsHap: fast and accurate read-based phasing. bioRxiv. 2016; 085050.
- Sedlazeck FJ, Rescheneder P, Smolka M, et al. Accurate detection of complex structural variations using single-molecule sequencing. Nat Methods. 2018;15(6):461-468.
For research use only. Not for use in diagnostic procedures. Unless otherwise agreed to in writing, IDT does not intend these products to be used in clinical applications and does not warrant their fitness or suitability for any clinical diagnostic use. Purchaser is solely responsible for all decisions regarding the use of these products and any associated regulatory or legal obligations. Doc ID: RUO22-1455_001