xGen™ SARS-CoV-2 Midnight Amplicon Panel
Multiplex PCR primers for amplicon sequencing
Panel for amplification of SARS-CoV-2 from research samples that can be used to understand the spread of the virus, identify variants, and helps enable a better understanding of COVID-19.
xGen™ NGS—made for COVID-19 research.
Ordering
- Amplifies the SARS-CoV-2 genome for sequencing using premixed multiplex PCR primers
- Provides data for understanding SARS-CoV-2 viral evolution and the epidemiology of COVID-19
- Data identifies SARS-CoV-2 variants, including alpha, beta, gamma, delta, mu, and omicron
- Panel generates 29 tiled amplicons, approximately 1200 bp in length
- Panel is optimized for high throughput applications or automated workflows
- Samples with Ct values <32 are recommended for complete genome sequencing coverage based on initial testing
Transform Your NGS Workflow with Automation
Looking to streamline your NGS workflows? Discover how automation can enhance efficiency and consistency in your lab with our NGS Automation solutions.
Product details
The Midnight Panel was created by Drs. Nikki Freed and Olin Silander of Massey University in New Zealand [1] and recently updated by Dr. John Tyson and Tracy Lee of the British Columbia Centre for Disease Control. This panel consists of 29 amplicons of approximately 1200 bp in length (Figure 1). The reduced number of amplicons allows for more consistent sequencing across the genome than would be achievable with a higher number of amplicons.
Figure 1. SARS-CoV-2 Midnight Amplicon Panel amplicons. The multiplex primer panel consists of two pools. The spacing is designed to tile across the entire SARS-CoV-2 genome, generating 29 amplicons approximately 1200 bp in length.
Since the primers only anneal to a total of 4.5% of the viral genome, the chance that viral mutations will disrupt primer annealing is lower. This results in a lowered chance of amplicon dropouts.
The Ct value for the amplicon sequencing reaction is an important metric since the higher the Ct, the lower the number of viral genomes in the sample. Samples with less viral genomic material pose difficulty for primer annealing and amplification.
In our experiment, contrived samples were prepared by mixing inactivated SARS-CoV-2 viral material into a negative background matrix of a negative nasopharyngeal (NP) swab in viral transport medium (VTM), with two samples per Ct value. Results show that when the SARS-CoV-2 Midnight Amplicon Panel and xGen DNA Library Prep Kit EZ are combined, the recommended Ct value is <32 which has been shown to sequence 97% of the SARS-CoV-2 genome with sufficient depth to identify variants (Table 1).
Table 1. SARS-CoV-2 Midnight Amplicon Panel and xGen DNA Library Prep Kit EZ metrics.
| Recommended Ct value | Coverage (%) | Contiguous amplicons (count) | Coordinates covered |
|---|---|---|---|
| <32 | 97% | 29 | 30–28,985 |
Workflow
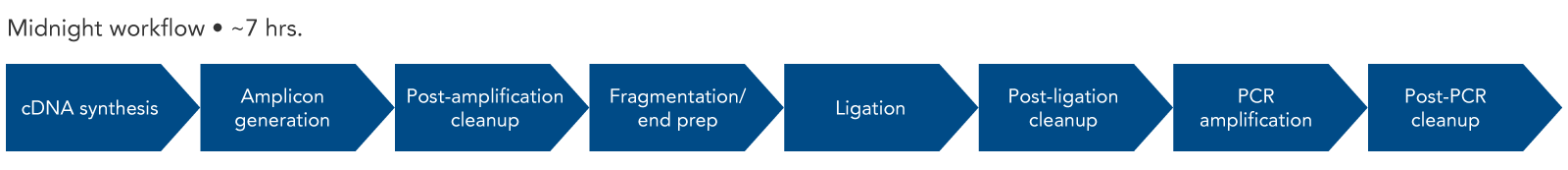
Since the length of the amplicons created by the SARS-CoV-2 Midnight 1200 Amplicon Panel are 1200 bp, Illumina® sequencing platforms are not compatible with this workflow. To use Illumina sequencing platforms, the SARS-CoV-2 amplicons generated with the Midnight panel can be fragmented using the xGen DNA Library Prep Kit EZ (with enzymatic DNA fragmentation). An overview of this workflow shows the steps necessary for library construction (Figure 2). IDT offers many of the reagents for the workflow, from the SARS-CoV-2 Midnight Panel and the components for library construction, to Nuclease-Free Water.
Figure 2. Expected experimental workflow for SARS-CoV-2 genome sequencing using the SARS-CoV-2 Midnight Amplicon Panel in combination with the xGen DNA Library Prep Kit EZ. By using the xGen DNA Library Prep Kit after the SARS-CoV-2 Midnight Amplicon Panel, the original 1200 bp fragments are converted into a library that is compatible with Illumina sequencers and chemistry.
For labs with access to Oxford Nanopore Sequencing instruments, the amplicons created using the SARS-CoV-2 Midnight Amplicon Panel can be prepared using one of the barcoding kits available directly from Oxford Nanopore (e.g., Rapid Barcoding Kit, SQK-RBK004).
Product data
As previously noted, the Ct value for the viral RNA in the sample is a critical metric to obtain usable data. IDT researchers used the SARS-CoV-2 Midnight Amplicon Panel in combination with the IDT xGen DNA Library Prep Kit EZ on samples obtained from nasopharyngeal swabs.
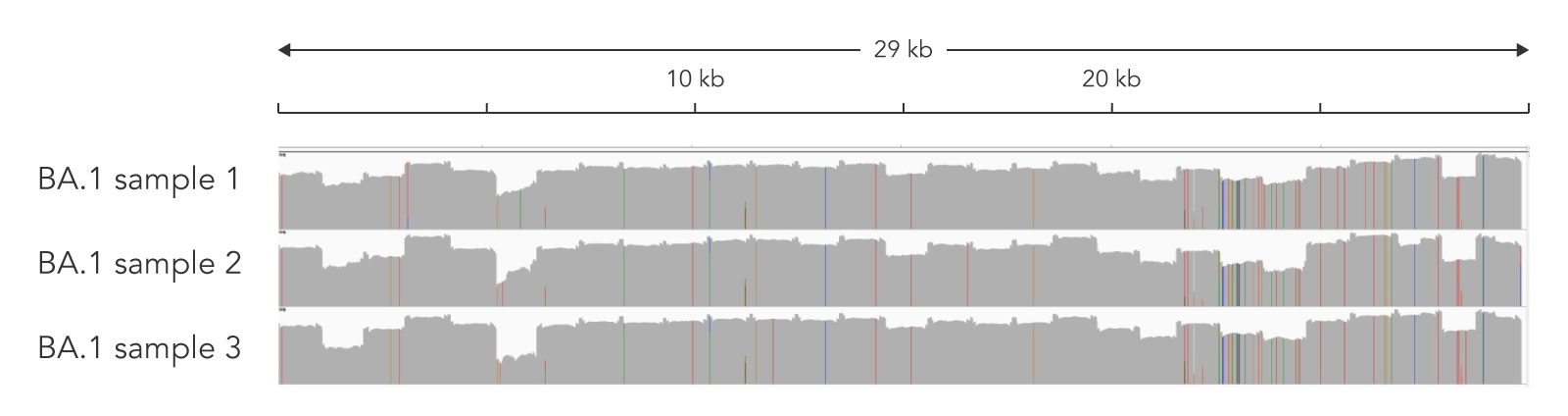
In a proof-of-concept test, sequencing libraries were first prepared according to the SARS-CoV-2 Midnight Amplicon Panel protocol for samples with varying Ct values. These libraries were then subjected to enzymatic fragmentation and library preparation according to the xGen DNA Library Prep Kit EZ recommendations (Figure 3).
Figure 3. Amplicon sequencing combining the SARS-CoV-2 Midnight Amplicon Panel and the xGen DNA Library Prep Kit EZ provided >99.5% coverage at >10X of the SARS-CoV-2 genome for Ct values 14−19. Data was generated from three independent samples of nasopharyngeal (NP) swab material with three different Ct values: 14.1, 19.9, and 15.3. Libraries were generated following the xGen DNA Library Prep Kit EZ recommendations. The resulting libraries were sequenced with MiSeq™ 300 cycle kit (2 x 150 reads) (Illumina). Samples were subsampled to 197,000 total reads. 100.0% of the reads were aligned to the viral genome and the coverage uniformity ranged from 81−91.9% with a mean coverage of ~950X. Pangolin analysis indicated all samples were Omicron strain (BA.1).
Resources
References
- Freed NE, Vlková M, et al. (2020) Rapid and inexpensive whole-genome sequencing of SARS-CoV-2 using 1200bp tiled amplicons and Oxford Nanopore Rapid Barcoding. Biol Methods Protoc 5(1):bpaa014.
- Freed N and Silander O (2020) nCoV-2019 sequencing protocol (RAPID barcoding, 1200 bp amplicon) v4 (protocols.io.bh7hj9j6). protocols.io.