Frequently asked questions
Our team has assembled a list of frequently asked questions to help you find answers quickly. Filter using one or more categories to focus on specific topics, or use the search bar to perform a text search.
Search all FAQs:
How do I use UMIs for error correction?
Depending on your sample type or experiment goals, you can choose to use UMIs or ignore them altogether.
The xGen™ cfDNA & FFPE Library Prep Kit Analysis Guidelines leads you through the recommended analysis pipeline using open-source tools starting with FASTQ files and resulting in variant calling.
As an overview, fixed UMI sequences, such as those used with the xGen cfDNA & FFPE Library Prep Kit, enable identification and correction of sequencing or PCR errors, even if they appear within the UMI sequence.
- Single-read families analysis: UMIs can also be used to correct errors in sequencing data at the same time as removing duplicate reads. For example, all reads with the same start-stop position and UMI can be grouped as a single-read family then collapsed. Rather than simply choosing the highest quality read, this method uses all reads within the single-read family to choose the most likely base at each position from beginning to end. This process yields a collapsed single-read family that can be used for variant calling. This approach is taken by the tools GroupReadsByUmi plus CallMolecularConsensusReads (fgbio).
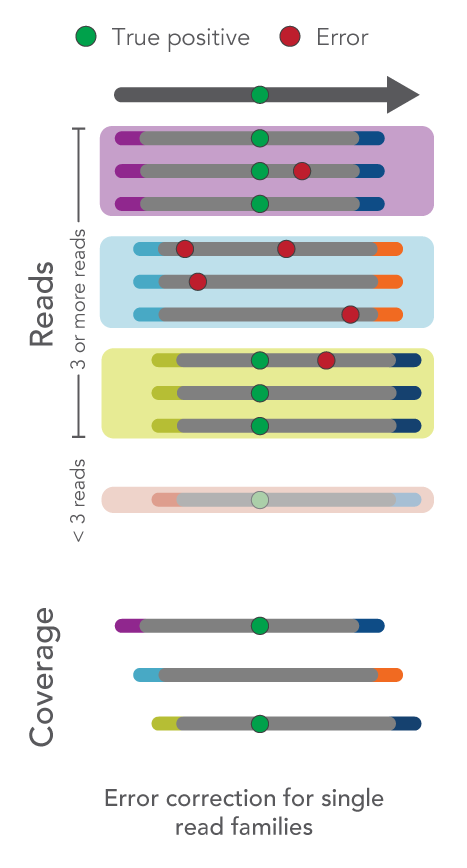
Figure 1. Schematic of error correction methods with UMIs.