PCR with RNase H (rhAmp™ PCR)
Overview
rhAmp PCR is RNase H-dependent PCR (rhPCR) that provides increased target affinity over traditional PCR through the use of RNase H2 and blocked primers.

What is rhAmp PCR?
rhAmp PCR is RNase H-dependent PCR (rhPCR) that provides increased target affinity over traditional PCR [1,2]. The difference between rhAmp PCR and traditional PCR lies in the use of an additional enzyme—RNase H2—and blocked primers.
How does it reduce primer-dimers?
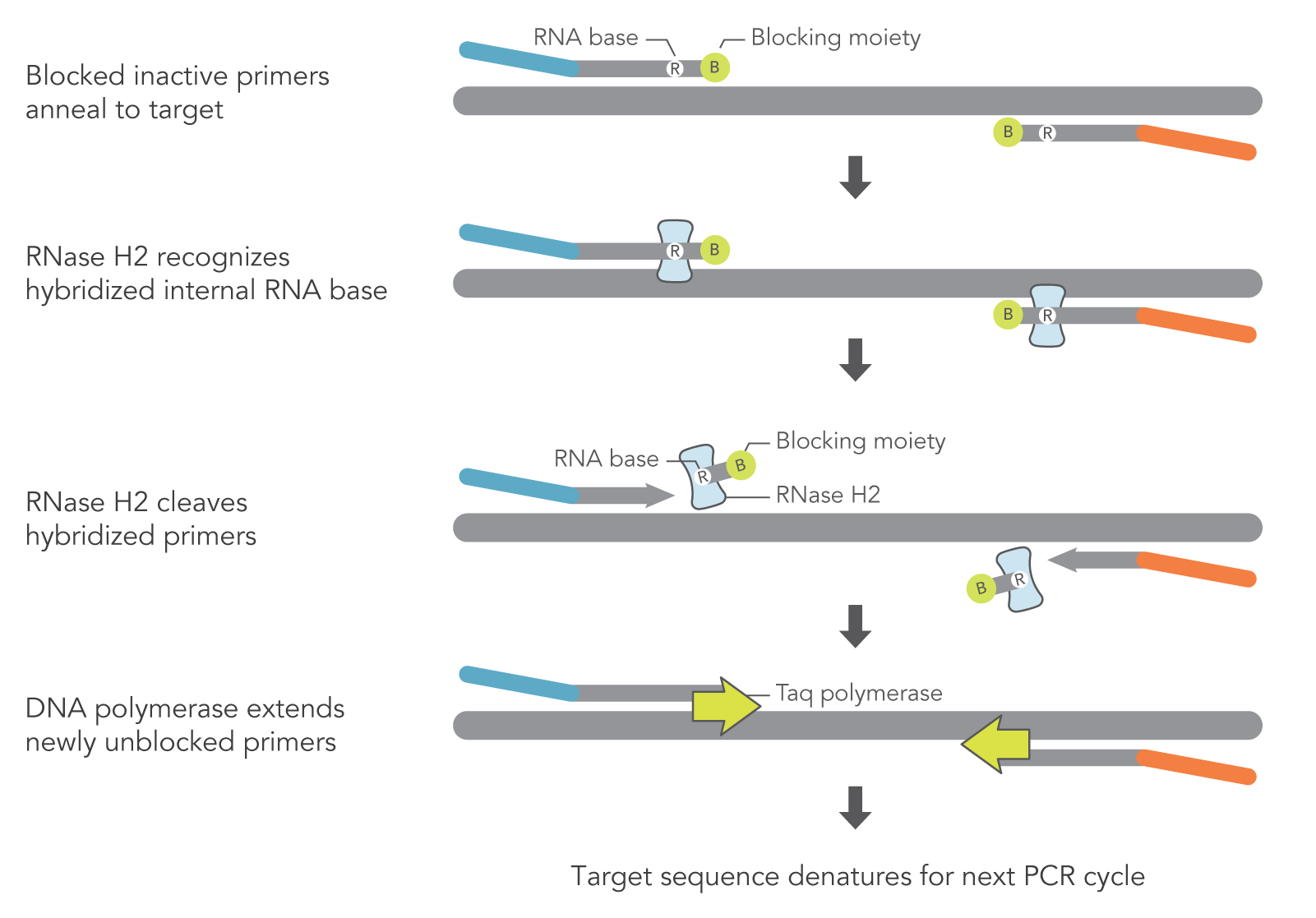
rhAmp technology relies heavily on the unique design of rhPrimers and the inherent characteristics of RNase H2. As opposed to DNA-only primers used in traditional PCR and qPCR, rhPrimers contain a single RNA base and a 3′ blocking group. RNase H2 must be added to the PCR reaction to remove the blocking moieties before extension by DNA polymerase can occur (Figure 1). In this way, the PCR reaction can result in more amplification of the target PCR fragments and less off-target amplification (Figure 2).
Who should use it?
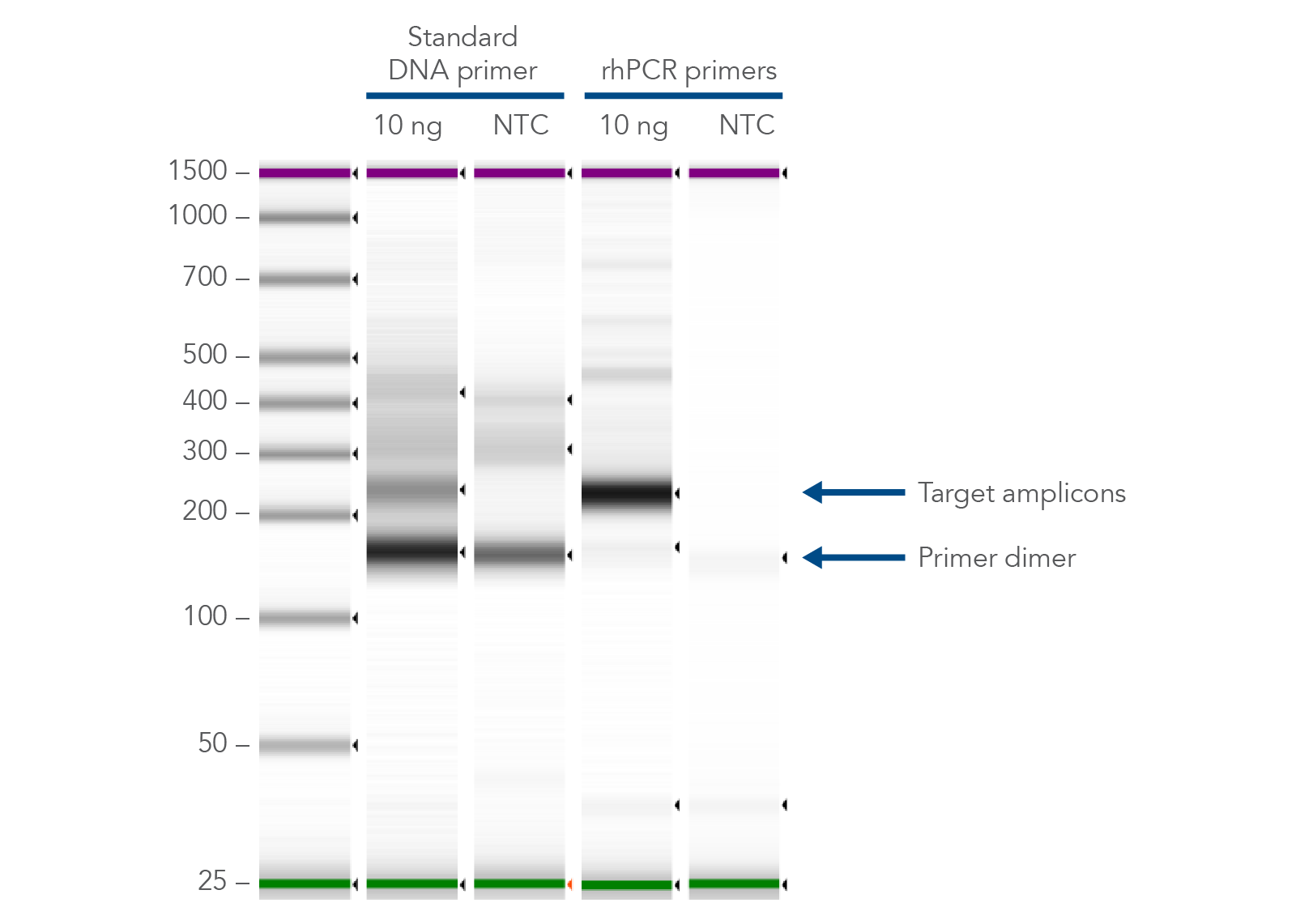
The primer-dimer reducing abilities of rhAmp technology make it ideal for genomics applications, like multiplexed amplicon sequencing and SNP genotyping. In addition, if you are investigating gene variants, splice variant discovery, or microbial identification, consider using rhAmp PCR to prepare your PCR assay. rhPrimers reduce primer-dimers and nonspecific amplification artifacts in multiplex applications, providing a practical alternative to regular PCR when undesirable amplification artifacts have become a problem (Figure 2).
Figure 1. Schematic representation of rhAmp PCR. DNA primers containing a single RNA residue and a 3′ blocking moiety (rhPrimers) are activated when cleaved by RNase H2 enzyme. Cleavage occurs on the 5′ side of the RNA base after primer hybridization to the target DNA. Because the primers can only be cleaved after they hybridize to the perfectly matched target sequence, primer dimers and other undesirable amplification products occur less often.
Figure 2. rhPrimers reduce primer dimers and nonspecific amplification artifacts in multiplex applications. In multiplex PCR amplification of 96 targets from human genomic DNA (NA12878, Coriell Institute), two sets of multiplex primers for the 96 assays (192 individual primers) were synthesized either as standard PCR primers or as rhPrimers. The results show tight control of primer-dimer artifacts, even in highly multiplexed assays. Total reaction volumes were 50 µL, with 10 ng of genomic DNA template or no template as controls. RNase H2 was present in all reactions but had no functional role in PCRs with standard primers.
Get started with genomics solutions based on rhAmp technology
Create your own PCR assay
References
- Dobosy JR, Rose SD, Beltz KR, et al. RNase H-dependent PCR (rhPCR): improved specificity and single nucleotide polymorphism detection using blocked cleavable primers. BMC Biotechnol. 2011;11:80.
- Beltz KT, D.; Wang, J. et al. A High-Performing and Cost-Effective SNP Genotyping Method Using rhPCR and Universal Reporters. Scientific Research. 2018;9(9).