Nickases vs. nucleases—How do the different Cas9 variants work?
The cleavage activity of the S. pyogenes Cas9 endonuclease is mediated by the coordinated functions of 2 catalytic domains, RuvC and HNH. These 2 domains function together to generate blunt-ended, double-strand breaks (DSBs) by cleaving opposite strands of double-stranded DNA (dsDNA) [1,2]. The RuvC domain cleaves the non-targeting strand—the strand containing the PAM sequence (protospacer adjacent motif, NGG for S. pyogenes Cas9), while the HNH domain cleaves the targeting strand—the strand complementary to the guide RNA (gRNA). A Cas9 nickase variant can be generated by alanine substitution at key catalytic residues within these domains: the RuvC mutant D10A produces a nick on the targeting strand while the HNH mutant H840A generates a nick on the non-targeting strand [3].
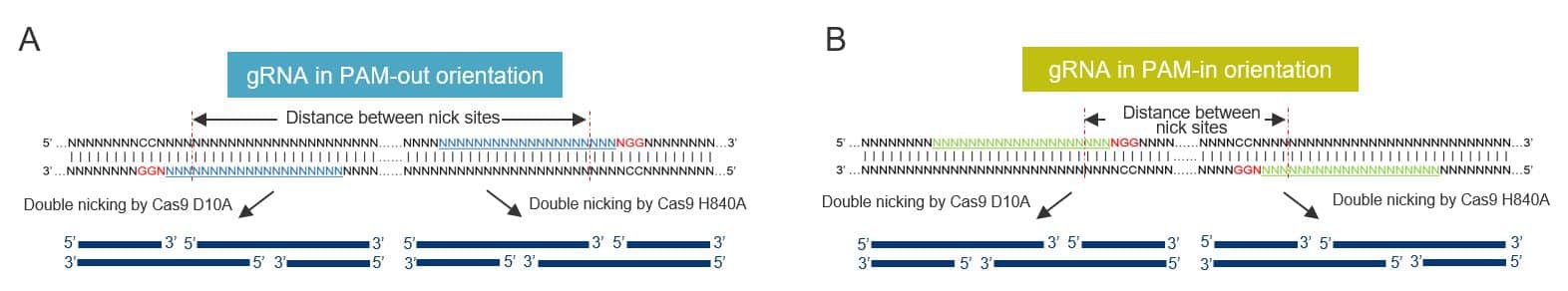
DSBs are known to be essential for efficient genome editing. In eukaryotic cells, neither single nor dual gRNAs targeting the same strand lead to efficient genome editing, because of a high-fidelity, base excision repair pathway. However, the combined use of 1 of the 2 Cas9 nickases with a pair of gRNAs targeting opposite DNA strands in close proximity generates a staggered DSB with overhangs (Figure 1).
Genome editing mediated by Cas9 nickases is inherently more complicated than a standard CRISPR-Cas9 experiment, given the requirement for 2 gRNAs to function simultaneously. Here, we briefly summarize a few considerations for successful nickase experimental design. In addition, we invite you to review the nickases section of our in-depth application note, Optimized methods for CRISPR-Cas9 homology-directed repair (HDR) for efficient, high-fidelity genome editing.
Top considerations for designing effective and efficient Cas9 nickase experiments
Design gRNA pairs with optimized spacing and orientation
A number of factors can impact the efficiency of cooperative nicking, such as type of overhang and hindrance effect between 2 adjacent Cas9 ribonucleoprotein (RNP) complexes. Through a systematic assessment of how gRNA designs affect indel formation by Cas9 D10A and H840A nickases, we found that high-efficiency editing was observed only when PAM sites faced the outside of the target region, a configuration known as “PAM-out” (Figure 1A). Furthermore, Cas9 D10A-mediated genome editing rates were improved when the 2 cleavage sites were 40–70 bp apart, while Cas9 H840A nickases favored a distance of 50–70 bp [4].
Design considerations for homology-directed repair (HDR) experiments mediated by Cas9 nickases
Researchers have been increasingly exploring the potential utility of Cas9 nickases for HDR-mediated sequence knock‑in. To support this use, IDT scientists sought to improve HDR efficiency by testing a variety of single-stranded oligonucleotide (ssODN) donor template design parameters, such as length of the homology arms, symmetry around insertion site, strand preference, and choice of nickase variant (D10A vs. H840A).
In agreement with a previous study [5], Cas9 D10A was more potent in mediating HDR than Cas9 H840A, despite having total editing efficiencies that are generally comparable [4]. When creating small insertions with single-stranded oligos (e.g., Alt-R™ HDR Donor Oligos), we have observed increased HDR rates with homology arm lengths of 30–60 bases (see application note). In addition, we found no consistent enhancement of HDR efficiency when the ssODN template was designed asymmetrically versus symmetrically.
Lastly, ssODN donor templates can be designed using the sequence of either strand of the template DNA. We recommend designing and testing templates complementary to both strands whenever possible, as strand preference differs between designs and is not always predictable.
Cas9 nickase experimental workflow
Based on the recommendations outlined above, a nickase experimental workflow can be established by using an Alt‑R Cas9 Nickase, Alt-R gRNA, and a synthetic donor template (optional, but required for HDR) to introduce nucleotide changes in a site-specific manner. Important steps for a successful nickase-mediated genome editing experiment include:
- Identify paired gRNAs close to the desired sequence change with ideal orientation (PAM-out) and distance.
- Test editing efficiency of gRNA pair candidates with nickases and select the pair with the highest editing efficiency.
- (Optional) Select D10A, rather than H840A, for nickase-mediated HDR experiments, although both variants can be potent in mediating gene disruptions.
- (Optional) Design a synthetic donor template and determine the optimal amount of donor template for your cells of interest.
- Design appropriate assays to confirm the desired genome editing outcomes in transfected cell lines.
Take away
While more involved than standard CRISPR-Cas9 experiments, the double-nicking strategy enabled by Cas9 nickase variants can be leveraged to reduce unwanted off-target editing, while maintaining high on-target efficiencies [6]. With a deliberately designed gRNA pair and careful optimization of experimental conditions, you can achieve precise and efficient genome editing with Alt-R Cas9 nickases.
References
- Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823.
- Jinek M, Chylinski K, Fonfara I, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821.
- Mali P, Aach J, Stranges PB, et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013;31(9):833–838.
- Schubert MS, Thommandru B, Woodley J, et al. Optimized design parameters for CRISPR Cas9 and Cas12a homology-directed repair. Sci Rep. 2021;11(1):19482.
- Bothmer A, Phadke T, Barrera LA, et al. Characterization of the interplay between DNA repair and CRISPR/Cas9-induced DNA lesions at an endogenous locus. Nat Commun. 2017;8:13905.
- Ran FA, Hsu PD, Lin CY, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154(6):1380–1389.
RUO22-1583_002