Generating a standard curve for absolute quantification
When performing quantitative PCR (qPCR), absolute quantification is usually accomplished by including artificial templates such as plasmids, oligonucleotides, or purified PCR products that have been accurately quantified by independent analysis. Standard curves plotted to known concentrations are then created by performing qPCR on serial dilutions of these templates.
Standard curves are useful for optimizing qPCR experiments, which is done by setting up qPCR reactions to amplify using different amounts of the same DNA sample. Ideally, you would need at least five data points over several orders of magnitude (5- to 10- fold dilutions)—to get a good standard curve.
Plasmids that have been sequenced are excellent for generating standard curves, but can be costly and time consuming to produce. Oligonucleotides and PCR products can be produced quickly, allowing greater flexibility when changing assays, but are limited in size. Additionally, PCR products may have unidentified sequence errors that will alter the efficiency calculation that is necessary during absolute quantification.
Using double-stranded gBlocks Gene Fragments as a standard curve template
gBlocks Gene Fragments, double-stranded, DNA fragments up to 3000 bp, can be used as an alternative to single-stranded oligonucleotides. Double-stranded DNA fragments function similarly to a double-stranded PCR product in applications, while offering the sequence flexibility of custom, chemically synthesized DNA. By including necessary sequence overlaps or restriction sites, gBlocks Gene Fragments are also ideally suited for isothermal assembly or cloning into any plasmid, providing a mechanism for production of additional high-fidelity template.
Generating multiple standard curves from a single template
The design flexibility of gBlocks Gene Fragments is useful when incorporating multiple control amplicon sequences into a single double-stranded construct. Using gBlocks fragments as multi-control templates lowers cost to a level that is comparable to ordering individual oligos on a per assay basis. It also provides some unique benefits when performing qPCR experiments in the lab:
- When performing multiplex experiments, combining control templates onto a single construct means less pipetting and, thus, less experimental variability. Each assay on that construct will have exactly the same amount of template available, providing for more accurate comparisons between those assays.
- For singleplex reactions, you need only make one set of dilutions. Those dilutions can then be used for all the assays represented on that construct. Again, this reduces the chances for pipetting error, and saves time diluting multiple, distinct templates.
When designing gBlocks Gene Fragments with multiple targets, some researchers choose to separate each sequence with several intervening T bases, as shown below. However, do not add more than 9 T bases between sequence elements, as this will interfere with the manufacture of your gBlocks fragment.

Use gBlocks Gene Fragments to detect contamination
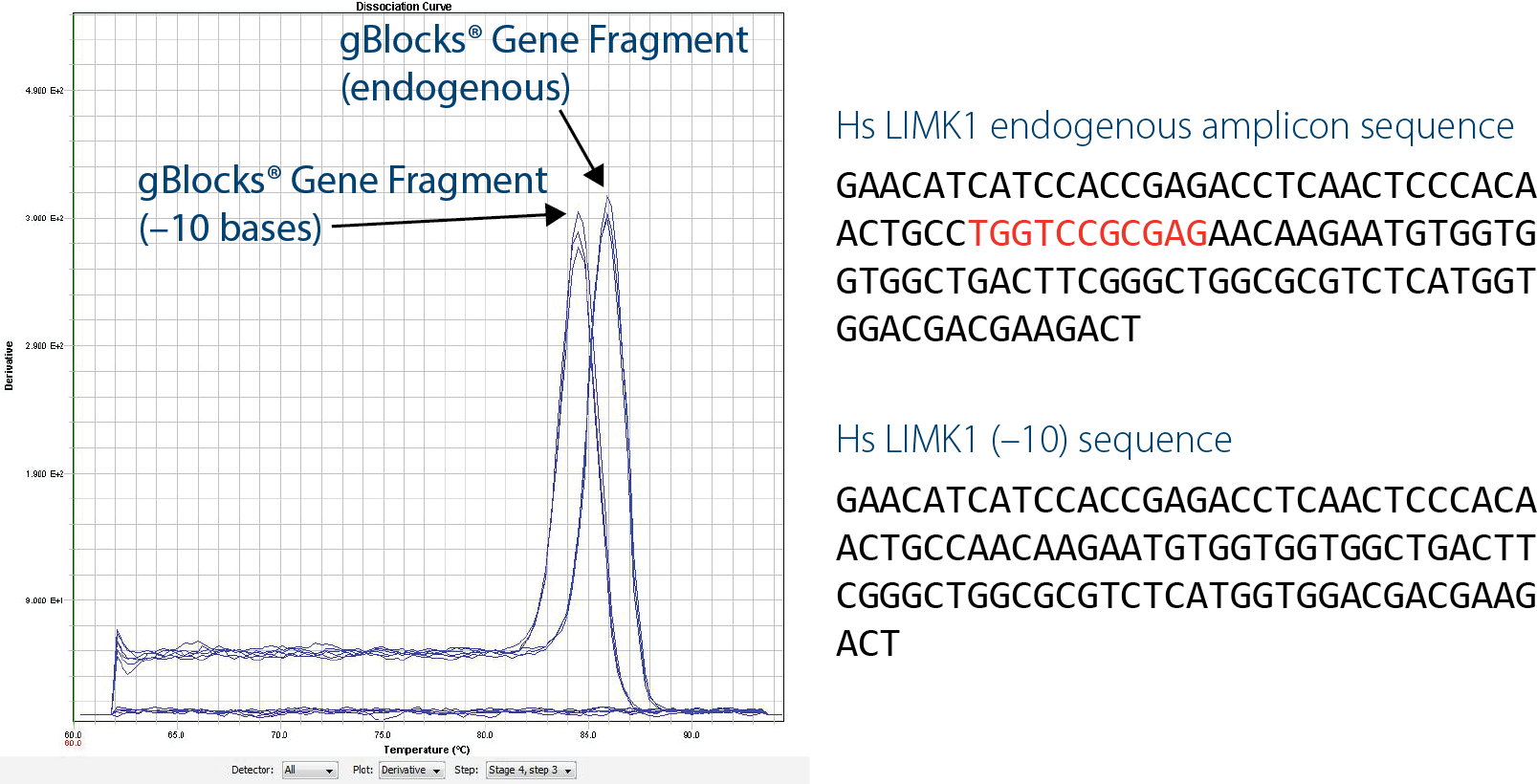
Another benefit of gBlocks Gene Fragments for qPCR is the ability to quickly generate artificial sequences that can be distinguished from wild-type sequences. An artificial construct that is 10 bp shorter than the wild-type sequence (LIMK1(–10)) can be distinguished by performing melt curve analysis using intercalating dye–based assays, such as with SYBR® Green. This is extremely beneficial if you have any concerns about possible contamination by wild-type DNA.
As an alternative, researchers can also consider a fully cloned, ready-to-use gene for generating qPCR controls.


 Processing
Processing

