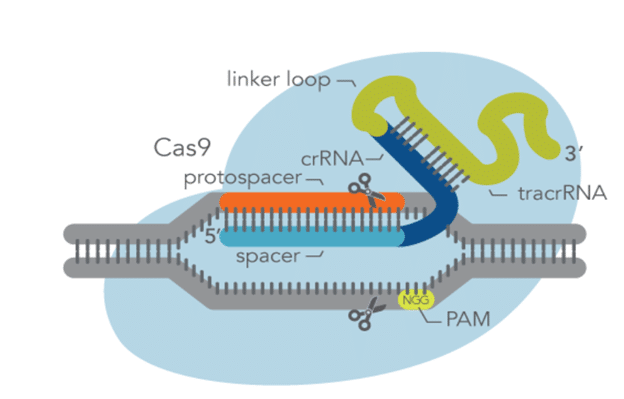
CRISPR genome editing technology utilizes a Cas enzyme and a guide RNA (gRNA) to generate mutations, gene knockouts, and knock-ins at a targeted genomic location. With Cas9, either a crRNA:tracrRNA duplex or an sgRNA can be used for CRISPR editing experiments. In either case, the Cas9 enzyme binds the gRNA to form a ribonucleoprotein (RNP). This allows the gRNA to then direct the RNP to the target site where the enzyme cleaves the DNA at a protospacer-adjacent motif (PAM). This PAM site is a specific DNA sequence where the Cas9 enzyme cleaves the DNA, making it important to properly design the gRNA so that it is targeting a DNA sequence of choice. In addition, the target sequence should be located close enough to a PAM sequence but also lead to high on-target potency while also resulting in reduced off-target activity.
What is an Alt-R™ CRISPR-Cas9 sgRNA?
Alt-R CRISPR-Cas9 sgRNA is an RNA oligonucleotide that contains both the target-specific crRNA region as well as the tracrRNA region that interacts with the Cas9 endonuclease all together in one molecule. It is 99 to 100 base pairs in length with 19 to 20 bases being the protospacer region and 80 bases the universal sgRNA region.
Cas9 sgRNA design factors
The two most important considerations when designing CRISPR sgRNAs for Cas9 are guide length and the sequence of the PAM site. The target sequence needs to be next to a PAM sequence of NGG, where N corresponds to any base, to be most effective and the optimal protospacer length for Cas9 is 20 base pairs. This is the target-specific spacer sequence and if this sequence is shorter than 20 nucleotides, then on-target editing efficiency decreases. The total length of a favorable sgRNA is 100 base pairs.
Getting started designing sgRNAs
To avoid manually selecting and evaluating these design criteria, sgRNA design can be done using the CRISPR guide RNA design tool from the IDT website. This allows you to look for predesigned sgRNA sequences or to design custom gRNA sequences as well as to check the gRNA sequence for both potential on- and off-target effects of the sequences you are considering.
- Search for Predesigned gRNAs - This option on the CRISPR sgRNA design tool allows you to select from a gRNA library of predesigned guide RNAs for 5 different species including human, mouse, rat, zebrafish, and C. elegans. All that is required is to enter the species of interest and either the gene symbol, accession number, or design id, and the number of designs you would like to generate. The tool then provides a list of designs that includes an on-target and off-target score for each gRNA. You can then select the gRNAs you would like to order directly from this list. It is recommended that you begin by testing 3 guides to find at least one or two that provide the highest editing efficiency.
- Design custom gRNAs - When selecting the custom guide RNA design option, you are able to enter a target sequence of your choice and are not limited by species. Either a FASTA sequence or a design ID can be used to input or select your target sequence. As with the predesigned gRNAs, an on-target and off-target score for each gRNA is also given but off-target analysis is only available for human, mouse, rat, zebrafish, or C. elegans. You can also select the gRNAs you would like to order directly from the list generated from this custom design. Just as with predesigned gRNAs, it is also recommended that you test at least 3 guides to find at least one or two that provide the highest editing efficiency.
- CRISPR-Cas9 gRNA Checker - Prior to ordering a gRNA sequence from a source other than the above tools, you can check the on- and off-target scores of any sequence using the CRISPR-Cas9 gRNA checker. This allows you to select the species as well as to enter a FASTA sequence to see what the predicted on- and off-target (options include human, mouse, rat, zebrafish, or C. elegans) scores are for the sequence of interest. It is important to only include the 20 base pair protospacer sequence that lies immediately 5’ to the PAM site for your target sequence but DO NOT include the PAM sequence itself.
To decipher the resulting on- and off-target scores you are aiming for high scores for both categories. A high on-target score indicates that you are likely to get high editing at the target site. A high off-target score suggests that there is not likely to be high off-target effects with this sgRNA. A combination of high on-target and high-off target scores should result in high on-target potency while also resulting in reduced off-target activity.
Ordering sgRNAs can be done directly from the CRISPR design tool or can also be done by entering the gRNA sequence here if you already know the exact sequence you want to order. Additional ordering options can also be found on our CRISPR Custom Guide RNAs page.
RUO23-2349_001