Considerations for better oligonucleotide design
Hybridization is a common step of many, if not most, molecular biology protocols. Examples of techniques that include nucleic acid hybridization are northern and Southern analysis, PCR/qPCR, cloning, in situ hybridization, array analysis, gene knockdown, and next generation sequencing (NGS). In this DECODED™ article, we learn from IDT scientist Dr Richard Owczarzy about oligonucleotide characteristics that impact melting temperature (Tm), a critical factor when planning experiments involving nucleic acid hybridization.
How do you calculate the Tm of DNA?
The criteria for hybridization are based on nucleic acid strand melting. Therefore, an understanding of melting temperature (Tm) provides information on when and how the DNA or RNA strands are going to hybridize and defines the rules for hybridization. It is very important to understand this process so that you can better design and optimize the oligos for your experiments. The Tm of an oligonucleotide duplex is the temperature at which half of the oligo molecules are single-stranded (and thus "melted") and half are double-stranded (i.e., annealed to its complementary strand). The accuracy of Tm calculations requires consideration of various physical properties of oligonucleotides, such as sequence length and composition, and accounts for experimental conditions, including the concentration of primers/probes, target oligo molecules, and salt/ionic strength. A convenient way to include all of these factors in your Tm calculations is to use a predesigned calculator, such as IDT's OligoAnalyzer™ Tool, available as part of our portfolio of online SciTools™ Web Tools.
Melting temperature (Tm) formula explained
Dr Richard Owczarzy (Ohv-char’-zee), a Principal Scientist at IDT and an extensively published expert on nucleic acid thermodynamics, confirms that many researchers are still under the illusion that they can use a simple formula to calculate Tm—multiplying the number of GC and AT bases by a given constant. “But this is not how Tm works,” says Dr Owczarzy. “Tm is not a constant value, but is dependent on the conditions of the experiment. Additional factors must be considered, such as oligo concentration and the environment in which hybridization will take place.” Important factors that affect Tm, including oligo concentration, ion concentration, sequence mismatch, and target strand choice, can all play a signification role in hybridization experiments.
Tm and oligonucleotide concentration
Tm varies with oligonucleotide concentration. While folding of a single oligonucleotide is concentration independent, the Tm is strongly influenced by oligo concentration when 2 or more nucleic acid strands interact. Oligo concentration alone can cause Tm to vary by ±10°C [2]. The molecule that is in excess determines Tm. For example, in PCR/qPCR, the target concentration is usually designed to be much lower than that of the oligo probe. In such situations, Tm is determined by the probe because it is in excess.
Does salt affect Tm?
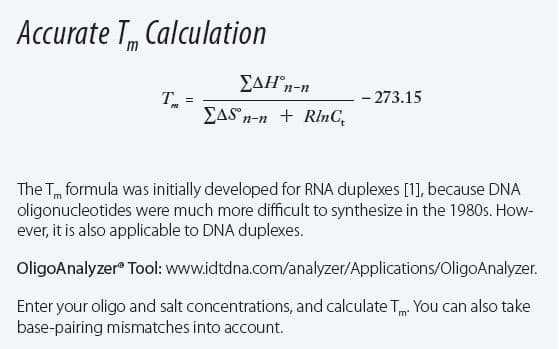
The concentrations of monovalent (sodium, potassium), divalent (magnesium), and polyvalent cations affect the stability of hybridized oligonucleotides [3,4]. Divalent cations have the biggest impact on Tm—changes in the millimolar range are significant. Increasing the concentration of monovalent cations, such as Na+, up to 1–2 M stabilizes oligos. However, these higher concentrations can significantly impact Tm. “A change from 20–30 mM Na+ to 1 M Na+ can cause oligonucleotide Tm to vary by as much as 20°C,” notes Dr Owczarzy. “We have worked through the calculations for sodium and magnesium ion concentrations and have come up with complicated models to predict Tm. This formula is used in the IDT SciTools programs to ensure your calculations are correct.” See the formula for Tm calculation in Figure 1 and explore the free online SciTools programs, such as the OligoAnalyzer Tool, available on the IDT website.
Only free Mg2+ in solution reacts with DNA; therefore, it is also important to consider the presence of any compounds that bind magnesium ions. For example, PCR requires deoxynucleoside triphosphates (dNTPs) in a mixture with short oligomer primers, probes, and longer nucleic acid targets. Magnesium ions bind to all of these components, thus decreasing the concentration of free Mg2+. As DNA synthesis proceeds during PCR, dNTPs are incorporated into the products and pyrophosphate is released. Pyrophosphate also binds Mg2+. Ideally, all of this should be considered when estimating the concentration of free Mg2+ in the hybridization solution.
Mismatches and single nucleotide polymorphisms (SNPs)
Mismatches between hybridizing oligos have a profound effect on Tm. The effect depends both on sequence context and solution composition (while salt is the main factor considered, other additives, like urea, DMSO, and even dyes such as SYBR® Green, can shift Tm). A single mismatch can cause Tm to vary between 1 and 18°C in PCR applications. The identity of the mismatch, its position in the sequence, and its context all impact the degree of the mismatch's effect. A-A and A-C are among the least stable mismatch pairs, causing the largest Tm variation, as compared to G-T, one of the more stable mismatch pairs. The context of the mismatch (e.g., whether a G-T mismatch is adjacent to an A-T or G-C base pair) also affects Tm. “It really requires a calculator to get an accurate prediction of Tm for one’s specific sequence, and there is no simple rule of thumb you can use without running the calculation,” says Dr Owczarzy.
SNPs that underlie primer and probe hybridization sites cause mismatches. "SNPs can affect PCR/qPCR experimental outcomes. There are a lot of SNPs out there, and their numbers are growing exponentially! Thus, we must design our PCR/qPCR assays intelligently, with SNPs in mind. It is important to check NCBI's dbSNP database to determine whether SNPs have been identified within your amplicon. Then you can attempt to design your primer and probe locations around those SNPs, as the mismatches they create could likely affect your PCR results,” warns Dr Owczarzy. "However, by positioning them towards the 5' end of the probe or primer sequences, thus, away from the 3' end, these effects can be minimized or even prevented."
Designing oligonucleotide probes for mismatch discrimination
When mismatch discrimination is the goal of an experiment, shorter probes will provide better mismatch identification. However, use of short probes, which will have a low Tm and thus require a low annealing temperature, can also reduce on-target hybridization. One way around this is to add stabilizing modifications such as locked nucleic acid residues (e.g., Affinity Plus™ modified bases) to raise probe Tm. Custom oligo modifications that affect Tm, including Affinity Plus locked nucleic acids, can now be directly entered into the OligoAnalyzer Tool for improved calculation accuracy.
Choice of target strand
Sense and antisense strands are not equal with regard to mismatch discrimination. PCR/qPCR produces complementary strands; therefore, you can choose the strand to which the probe will bind. Thus, if you aim to identify a single base change in one strand, you can choose to design the primers and/or probe to match the corresponding change of the complementary base in the other strand. The probe sequence used to bind to the sense strand will form a different type of mismatch than the sequence required to bind to the antisense strand. It may, therefore, be useful to calculate Tm for both scenarios. The free, online IDT OligoAnalyzer Tool allows researchers to calculate Tm for mismatched sequences. You can also use the program to determine probe locations that help enable better mismatch discrimination.
References
- Freier SM, Kierzek R, Jaeger JA, et al. Improved free-energy parameters for predictions of RNA duplex stability. Proc Natl Acad Sci, USA. 1989;83(24):9373–9377.
- Ozel AB, Srivannavit O, Rouillard JM, Gulari E. Target Concentration Dependence of DNA Melting Temperature on Oligonucleotide Microarrays. Biotechnol Prog. 2012;28(2):556–66.
- Owczarzy R, Yong Y, Moreira BG, et al. Effects of sodium ions on DNA duplex oligomers: improved predictions of melting temperatures. Biochemistry. 2004;43(12):3537–3554.
- Owczarzy R, Moreira BG, Yong Y, Behlke MA, Walder JA. Predicting stability of DNA duplexes in solutions containing magnesium and monovalent cations. Biochemistry. 2008;47(19):5336–53.
For research use only. Not for use in diagnostic procedures. Unless otherwise agreed to in writing, IDT does not intend these products to be used in clinical applications and does not warrant their fitness or suitability for any clinical diagnostic use. Purchaser is solely responsible for all decisions regarding the use of these products and any associated regulatory or legal obligations. RUO22-1656_001