CRISPR Off-target Analysis Services
Advanced, end-to-end workflow with orthogonality
For customers seeking to ensure the safety and efficacy of their CRISPR-based therapeutic developments through off-target analysis, our comprehensive end-to-end services encompass assays from off-target nomination to confirmation, with outputs specifically tailored for IND filings.
Rapidly move from the lab to life-changing advances.
Overview
CRISPR Off-target Analysis Services
Screening for off-target effects (OTE) is crucial to ensure the safety and efficacy of CRISPR-based therapies. By identifying and minimizing these unintended edits, researchers can reduce the risk of adverse outcomes and improve the precision of genome editing. This is particularly important in clinical applications, where unintended genetic modifications could have serious health implications for patients. By leveraging our enhanced OTE services, researchers and cell and gene therapy (CGT) developers can gain a deeper understanding of the off-target cleavage activities associated with CRISPR-Cas nucleases. This knowledge is crucial for developing safer and more effective genome editing therapies, ultimately advancing the field of genetic medicine and improving patient outcomes. Request a consultation today.
Why choose Integrated DNA Technologies' off-target editing analysis services?
- Comprehensive end-to-end assessments: Utilize next-generation sequencing-based off-target analysis technology, UNCOVERseq is based on GUIDE-seq™, alongside the award-winning rhAmpSeq™ CRISPR Analysis System for your pre-clinical therapeutic development.
- Robust performance, rigorous analytical and process standards: Experience highly reproducible assays with analytical sensitivities driven by empirical data for both nomination and confirmation assays.
- Expert regulatory support: Navigate complex FDA and EMA guidance for IND-enabling projects with our specialized regulatory support, tailored to meet the stringent requirements of your therapeutic development.
- Comprehensive reporting: Receive in-depth reports on off-target nomination and confirmation, featuring site prioritization and recommended confirmation panels, providing you with actionable insights for your research or therapy development.
- Dedicated CRISPR team: Partner with our team of CRISPR experts, committed to your project's success from inception to completion, ensuring personalized support and expert guidance throughout the entire process.
Services
Ensuring the safety and efficacy of your cell and gene therapy program earlier in the discovery process is more critical than ever. Outsource your CRISPR off-target analysis to enable reliable gene editing safety assessments for IND enabling studies based on latest guidance from the regulatory bodies.
End-to-end CRISPR off-target analysis from discovery to IND
Our comprehensive services include assays for both nomination and confirmation, utilizing orthogonal methods from discovery through preclinical stages and IND fillings.
| Assessment stage | Use case | Assay description |
|---|---|---|
| Off-target nomination |
|
|
| Off-target confirmation |
|
|
Services aligned with guidance from FDA
Services for both nomination and confirmation assays are aligned with FDA guidance.
| FDA and other agencies | Nomination stage | Confirmation stage |
|---|---|---|
| Utilize multiple complementary assessment methods | Yes | |
| Detect off-target effects with high analytical sensitivity | Yes | Yes |
| Analyze human cell types from multiple donors | Yes | Yes |
| Detail the biological impact of off-target effects | Yes | Yes |
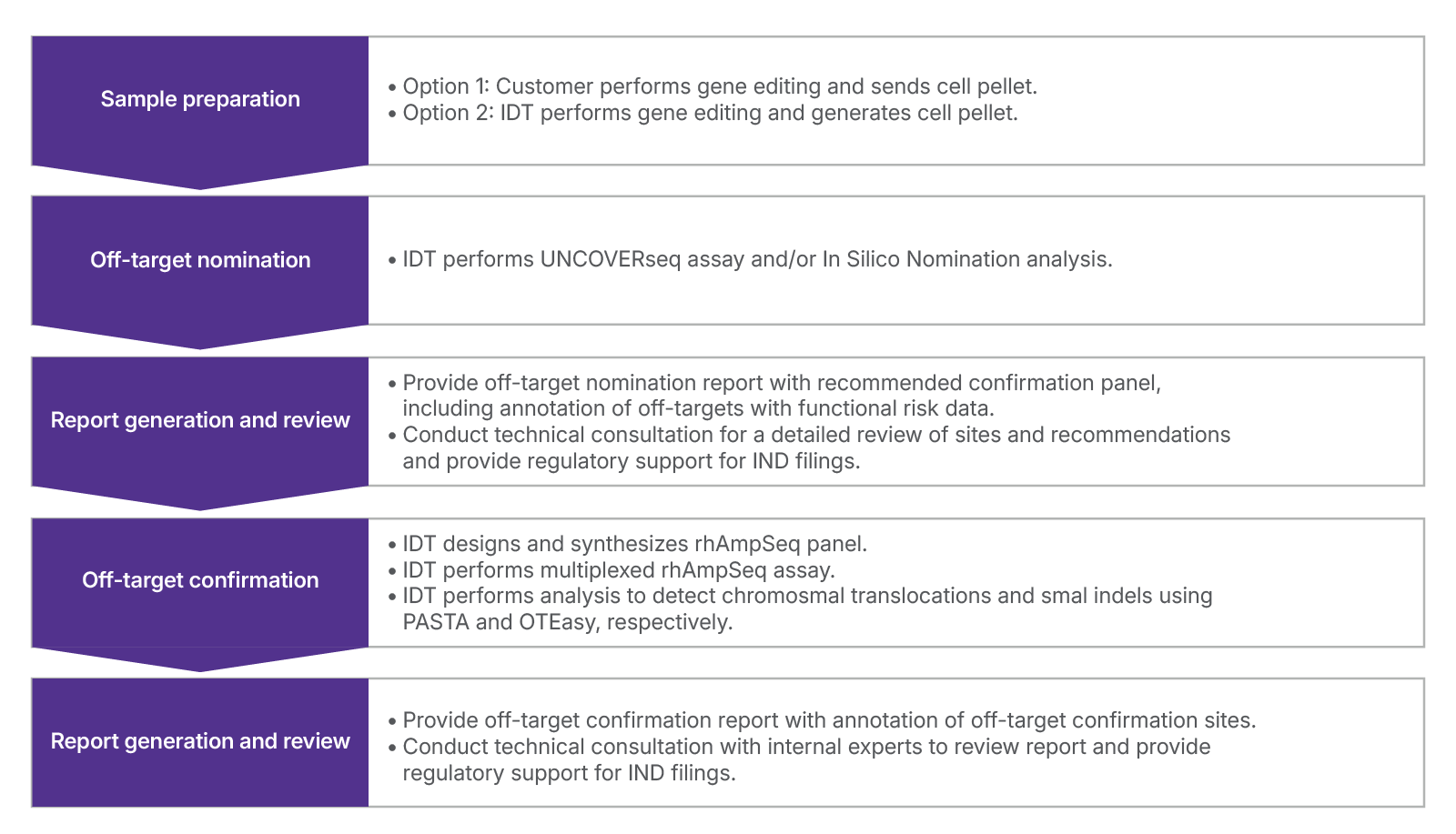
Our end-to-end process:
Leverage our services from nomination to confirmation, or select individual services tailored to your specific needs. For greater flexibility, you can either send us your samples or have IDT generate the samples using in-house nomination model systems.
Nomination assays
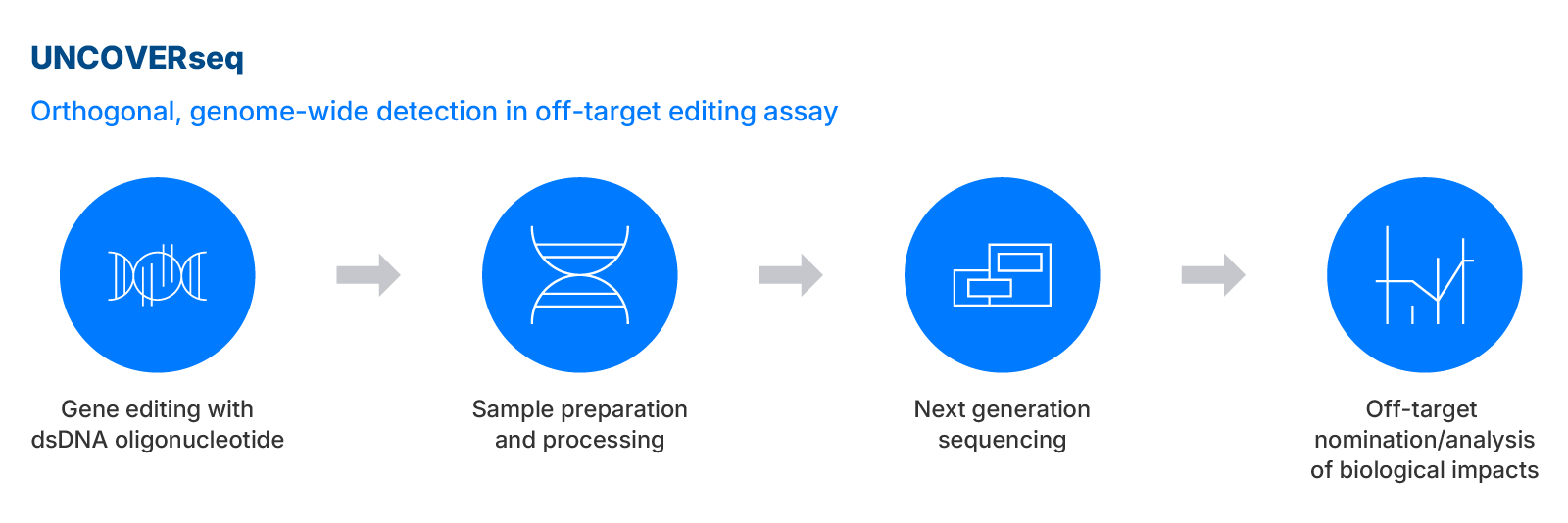
UNCOVERseq
IDT’s UNCOVERseq is a next-generation sequencing method based on the cell-based GUIDE-seq™ method for off-target nomination. UNCOVERseq stands for Unbiased Nomination of CRISPR Off-target Variants with Enhanced RhPCR.
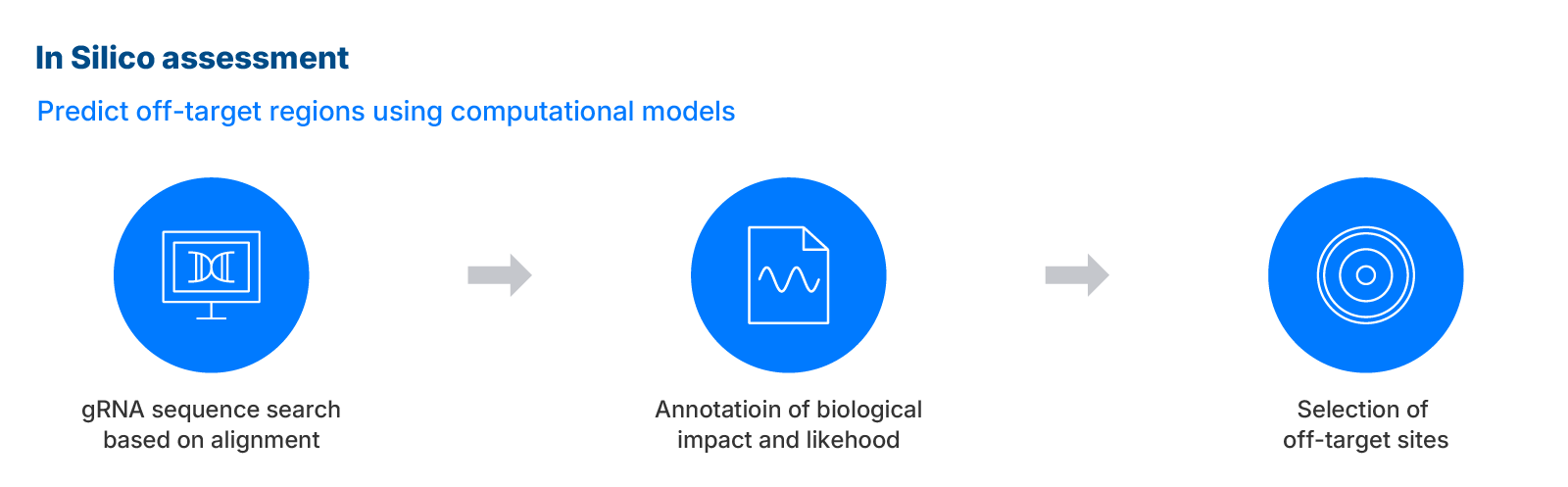
In silico assessment
IDT's In silico assessment leverages computational methods to nominate potential off-target regions for CRISPR edits. This approach, combined with UNCOVERseq, ensures thorough annotation and prioritization of off-target sites, enhancing the accuracy and reliability of genotoxicity evaluations.
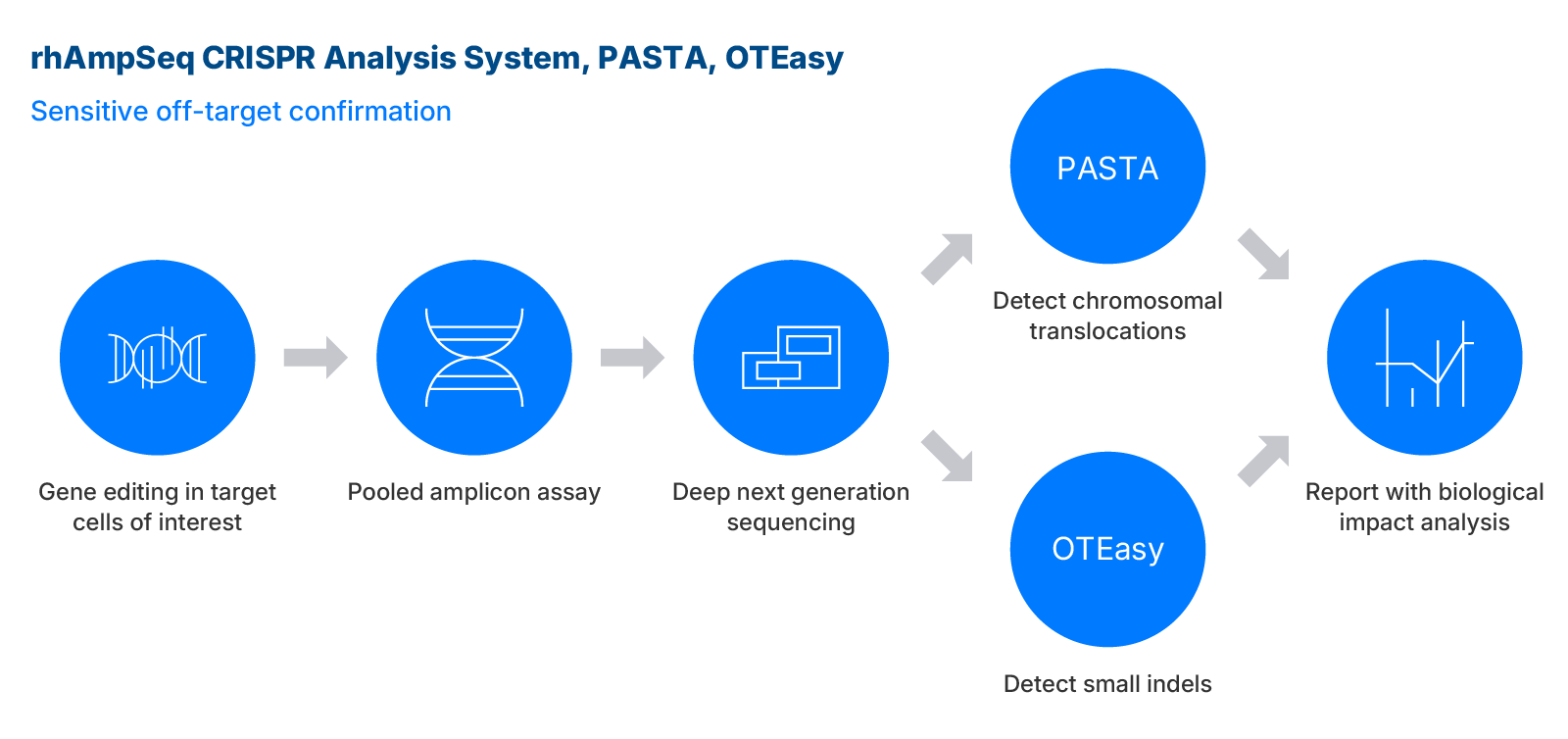
Confirmation assays
rhAmpSeq CRISPR analysis assay and tools
The award winning rhAmpSeq™ CRISPR Analysis System can be used to identify small indels, base editing, and chromosomal aberrations using high-throughput amplicon sequencing coupled with IDT's analysis workflows.
Resources
Related products
cGMP25-3559_001
GUIDE-seq™ is owned by Maxcyte®, Inc.
FOR INFORMATIONAL PURPOSES ONLY. The results provided are for informational use only and should not be used as the sole basis for any critical decision making. The results provided herein are based on assay procedures which have not undergone full validation; formal design and development activities are on-going. Purchasers are responsible for all decisions regarding the use of this information.

