PrimeTime™ Gene Expression Master Mix for qPCR
Obtain high-quality results with this thermostable probe-based qPCR master mix
PrimeTime Gene Expression Master Mix is a 2X solution containing a hot-start antibody, Taq polymerase, and other components needed for probe-based qPCR in two-step RT-PCR experiments. Reference dye is included separately.
Ordering
- High efficiency qPCR with fast or standard cycling
- Use one mix for single-plex or multiplex reactions
- Rely on benchtop stability to obtain consistent results from overnight experiments
Need 500 mL or more? Contact us for discounted pricing.
Product details
PrimeTime Gene Expression Master Mix has been formulated for probe-based qPCR assays such as PrimeTime predesigned sequences for gene expression analysis. PrimeTime predesigned sequences will result in >90% efficiency, or we will replace with an alternative design free of charge. The PrimeTime Gene Expression Master Mix is also compatible with other primers and probes.
Each order includes a 2X master mix solution (antibody-mediated, hot-start DNA polymerase; dNTPs; MgCl2; enhancers; and stabilizers) and a separate reference dye stock solution.
PrimeTime Gene Expression Master Mix is shipped at ambient temperature. Elimination of shipping on dry ice saves on shipping costs, minimizes shipping delays, and benefits the environment. To view our testing that shows ambient shipping does not impact the functionality of the PrimeTime Gene Expression Master Mix, see our white paper.
For a general estimate, the following table provides the number of reactions (for 20 µL reaction volumes) for each order size:
| Product | Catalog # | Unit size | Reactions |
|---|---|---|---|
| PrimeTime Gene Expression Master Mix, 1 mL | 1055770 | 1 x 1 mL | 100 x 20 µL |
| PrimeTime Gene Expression Master Mix, 5 mL | 1055772 | 1 x 5 mL | 500 x 20 µL |
| PrimeTime Gene Expression Master Mix, 25 mL | 1055771 | 5 x 5 mL | 2500 x 20 µL |
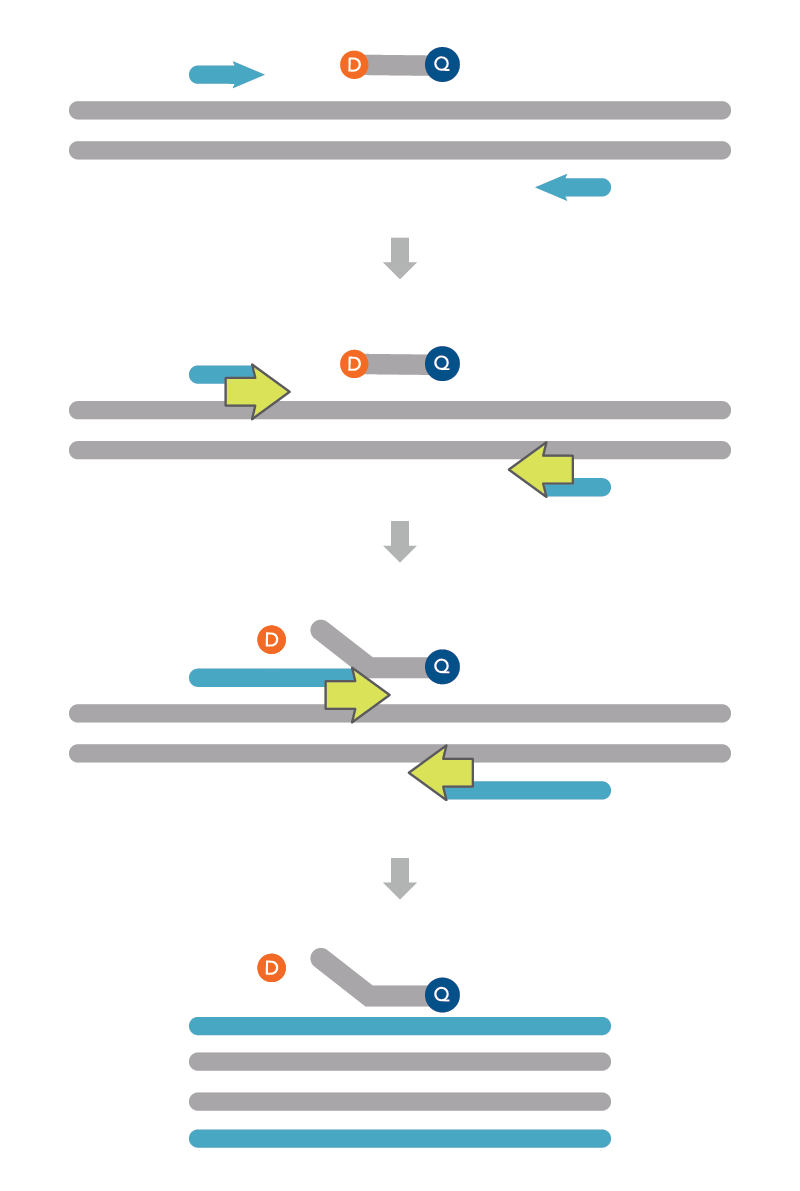
Overview of 5′ nuclease assays
Step 1—The primers and probe hybridize in a sequence-dependent manner to the complementary DNA strand. Because the probe is intact, the fluorophore and quencher are in close proximity and the quencher absorbs fluorescence emitted by the fluorophore.
Step 2—The polymerase extends from the primers and begins DNA synthesis.
Step 3—The polymerase reaches the probe and the exonuclease activity of the polymerase cleaves the hybridized probe. As a result of cleavage, the fluorophore is separated from the quencher and fluoresces.
Step 4—The fluorescence is detected by the real time instrument.
These steps are repeated for each PCR cycle and allow detection of specific products. When using intercalating dyes, such as SYBR® Green I, primer-dimers and off-target hybridization products will also contribute to fluorescence. In contrast, the 5′ nuclease assay results in more on-target binding, and fluorescence will only be observed for the DNA sequence to which the probe and primers hybridize.
Product data
Compatible with PrimeTime qPCR Assays (and assays from other suppliers)
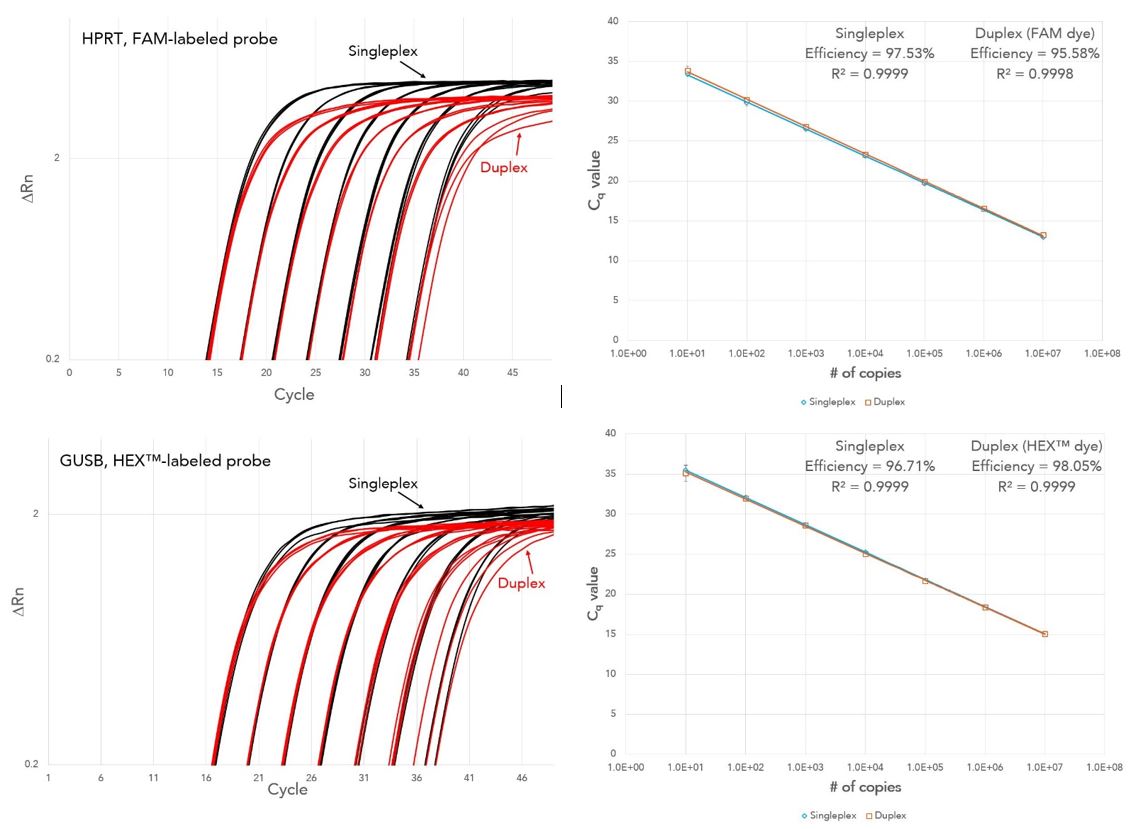
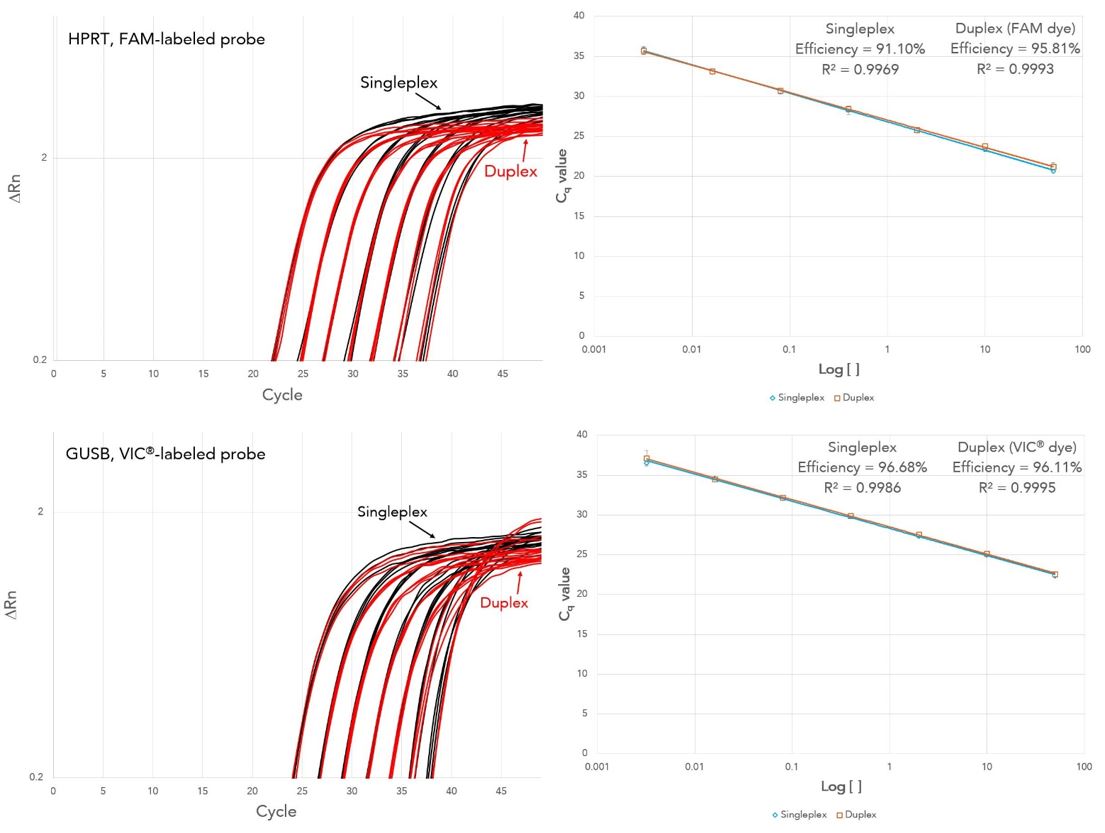
PrimeTime Gene Expression Master Mix delivers reliable results with single-plex and duplex amplification using qPCR assays from IDT or other suppliers. All assays exhibited 90−110% PCR efficiency with R2 >0.99 (Figures 2 and 3).
Figure 2. Reliable results from single-plex (black) and duplex (red) qPCR using PrimeTime qPCR Assays. PCRs, containing PrimeTime HPRT and/or GUSB qPCR Assays, PrimeTime Gene Expression Master Mix with reference dye, and gBlocks Gene Fragments (101–107copies) as template, were run on a 7900HT Real-Time PCR System (Thermo Fisher Scientific). HPRT Assay ID: Hs.PT.58v.45621572; GUSB Assay ID: Hs.PT.58v.27737538 (n = 3).
Figure 3. Reliable results from single-plex (black) and duplex (red) qPCR using inventoried HPRT and GUSB qPCR Assays (Vendor A). PCRs, containing inventoried HPRT and/or GUSB qPCR assays from Vendor A, PrimeTime Gene Expression Master Mix with reference dye, and cDNA (0.0032−50 ng) as template, were run on a 7900HT Real-Time PCR System [(Thermo Fisher Scientific), n = 3].
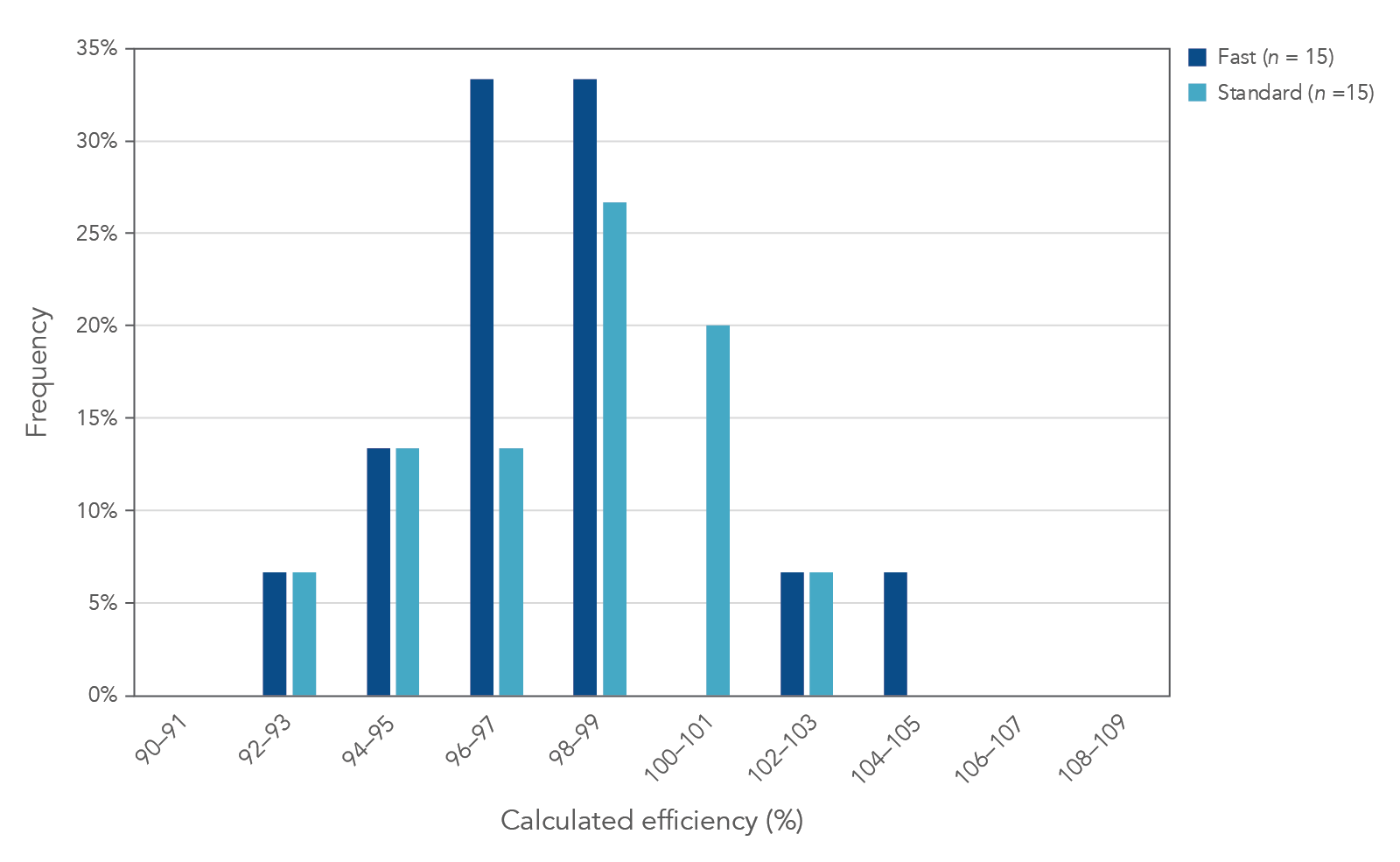
Standard or fast cycling conditions
PrimeTime Gene Expression Master Mix performs well under standard or fast cycling conditions (Figures 4 and 5) on a variety of real-time qPCR instruments, including the 7900HT Real-Time (Thermo Fisher Scientific), QuantStudio™ 7 Flex (Thermo Fisher Scientific), CFX384 (BioRad), and LightCycler®480 (Roche) qPCR systems.
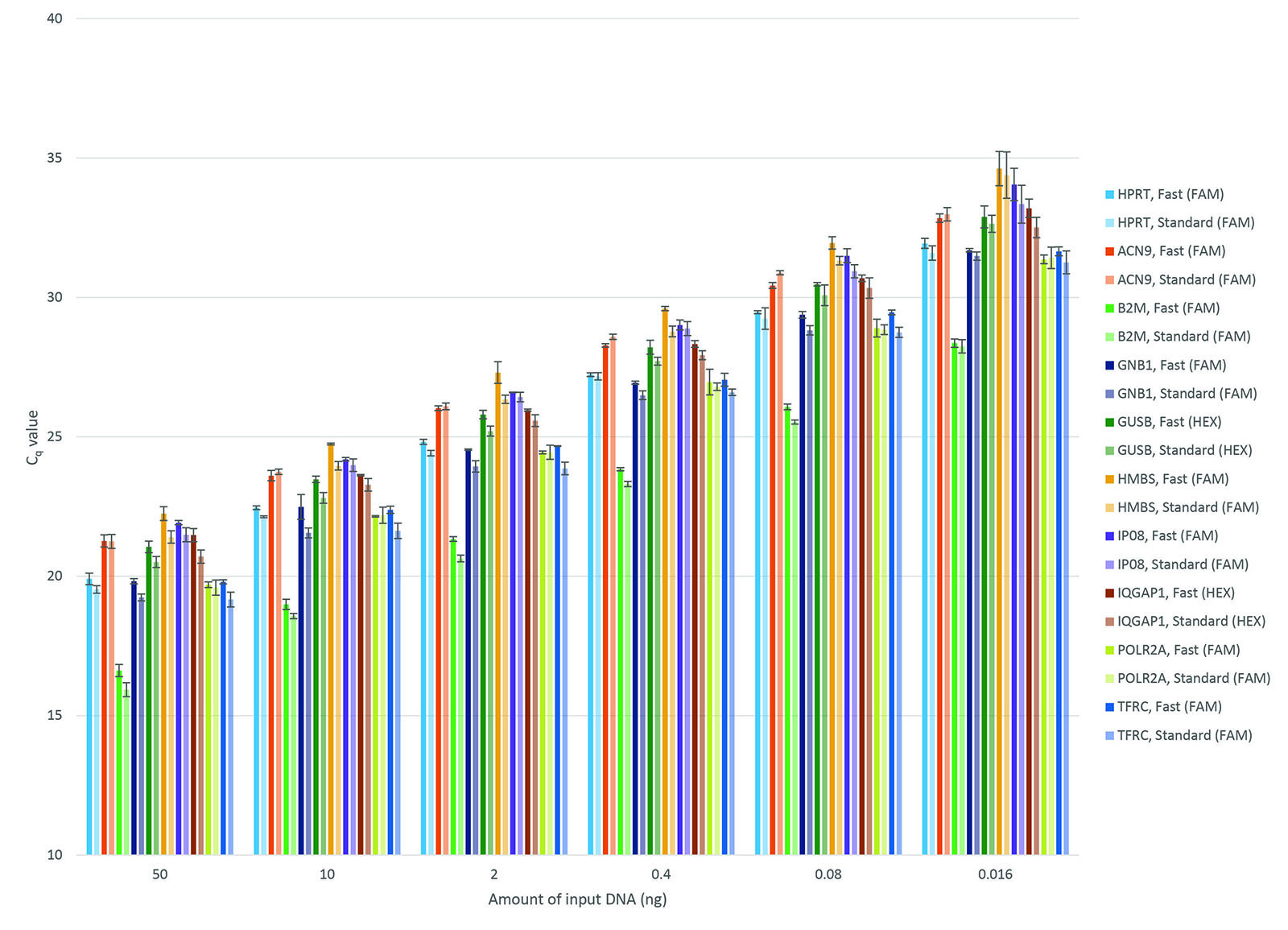
Figure 4. High PCR efficiency under standard or fast cycling conditions. PCRs consisting of PrimeTime qPCR Assays, PrimeTime Gene Expression Master Mix, reference dye, and template were run on a 7900HT Real-Time PCR System (Thermo Fisher Scientific) using standard or fast cycling conditions. Standard cycling conditions: 3 min. 95°C; 49 x (15 sec. 95°C; 1 min. 60°). Fast cycling conditions: 3 min. 95°C; 49 x (5 sec. 95°C; 30 sec. 60°). This histogram shows the calculated PCR efficiency of 15 assays run under standard (light blue) or fast (dark blue) cycling conditions using either diluted cDNA (0.016−50 ng) or gBlocks Gene Fragments (101−107 copies) as template. All assays exhibited 90−110% PCR efficiency with R2 >0.99.
Figure 5. Consistent Cq values under standard or fast cycling conditions. PCRs consisting of PrimeTime qPCR Assays, PrimeTime Gene Expression Master Mix, reference dye, and dilutions of cDNA (0.016−50 ng) were run on a 7900HT Real-Time PCR System (Thermo Fisher Scientific) using standard or fast cycling conditions. Standard cycling conditions: 3 min. 95°C; 49 X (15 sec. 95°C; 1 min. 60°). Fast cycling conditions: 3 min. 95°C; 49 X (5 sec. 95°C; 30 sec. 60°). At each concentration of cDNA, the difference in Cq values determined using standard or fast cycling conditions was <1 (n = 3). Error bars represent standard deviation.
Lot-to-lot consistency (0–24 hr at room temperature)
You can expect consistent results even when using different lots of PrimeTime Gene Expression Master Mix. This master mix has also shown temperature stability after 24 hours at room temperature, making it ideal for high-throughput applications or overnight experiments.
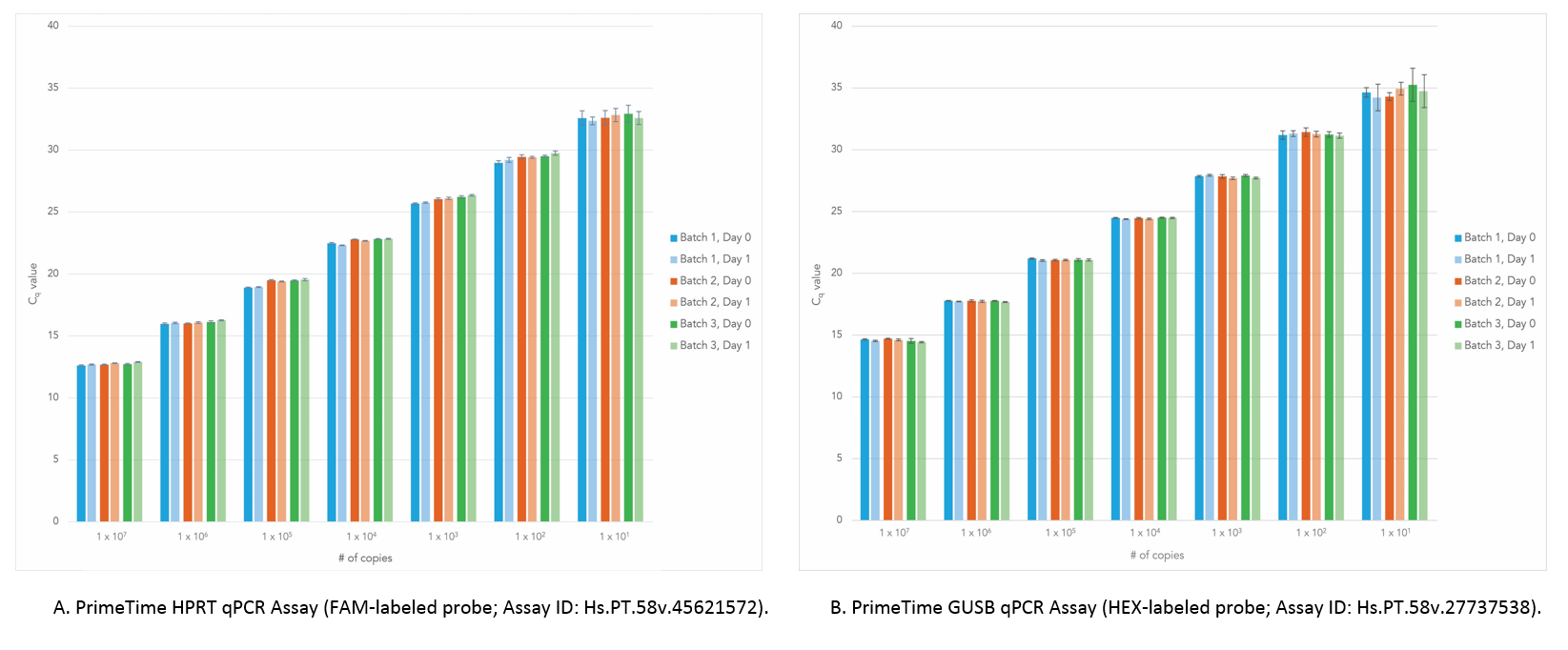
Figure 6. Dependable results across different batches of PrimeTime Gene Expression Master Mix, even after 24 hours at room temperature. PCR mixes consisting of PrimeTime qPCR Assays, PrimeTime Gene Expression Master Mix, reference dye, and varying amounts of gBlocks Gene Fragments (n = 8) were assembled. A JANUS® automated, liquid-handling workstation (PerkinElmer) was used to load the reactions into PCR plates, which were run immediately (Day 0) or after 24 hours (Day 1) on a 7900HT Real-Time PCR System (Thermo Fisher Scientific). When comparing the results from Day 1 with those from Day 0, the Cq values for template levels >10 copies were <0.5. (A) Results using the PrimeTime HPRT qPCR Assay. (B) Results using the PrimeTime GUSB qPCR Assay. Error bars represent standard deviation.
Temperature stability
In addition to consistent results after extended periods at room temperature, PrimeTime Gene Expression Master Mix has shown impressive temperature stability, with no loss of amplification or degradation of components, even after temperature stress tests (4 or 8 hours at 55°C, Figure 7). For additional stability data (e.g., up to 7 days at 50°C; up to 20 freeze-thaw cycles), see the white paper.
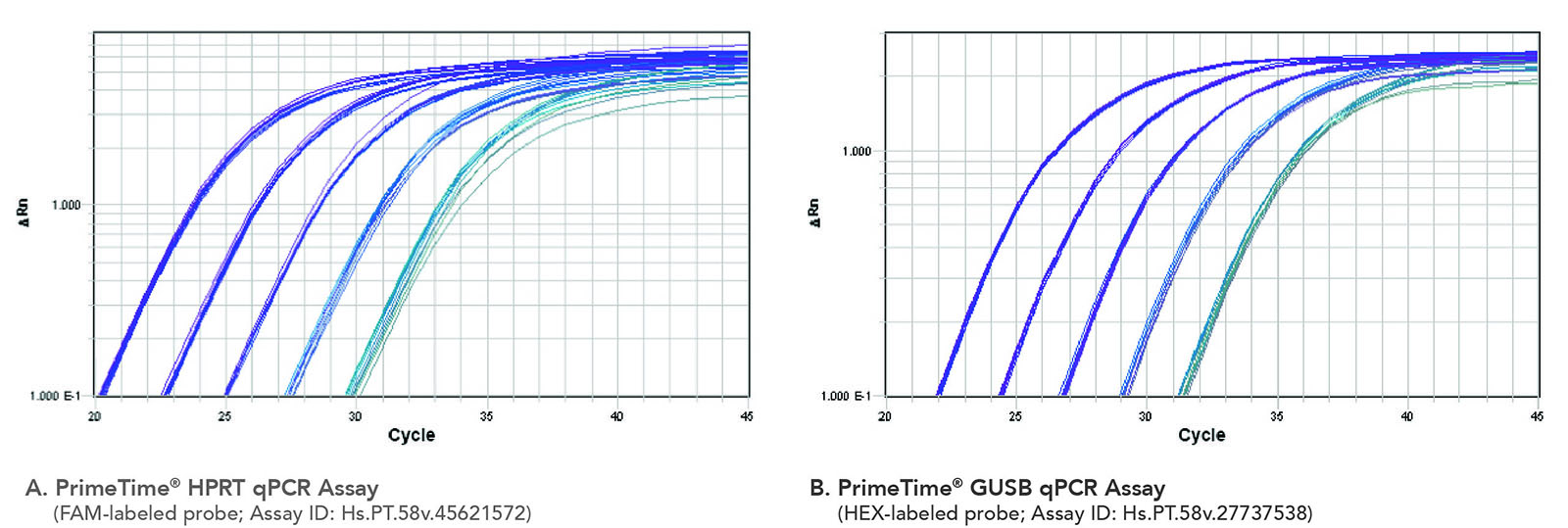
Figure 7. Consistent amplification after PrimeTime Gene Expression Master Mix was heated at 55°C for 4 or 8 hr. PrimeTime Gene Expression Master Mix was not heated or heated at 55°C (4 or 8 hr) before use in PCR with a PrimeTime qPCR Assay, reference dye, and varying amounts of cDNA (0.08−50 ng). An overlay of the amplification plots for the PCRs using not heated and heated master mix shows no effect on the PCR amplification curves. (A) Results using the PrimeTime HPRT qPCR Assay. (B) Results using the PrimeTime GUSB qPCR Assay (n = 3).
Services
Bulk orders
For orders >500 mL, contact us.
GMP* or OEM
If you require probes that are ISO 13485:2016 certified, or if you are interested in our third-party manufacturing services, please click here.
*GMP refers to products manufactured under ISO 13485:2016 QMC. Purchaser is solely responsible for all decisions regarding the use of these products and any associated regulatory or legal obligations for their legal marketing.
Resources
Frequently asked questions
How long can I leave my qPCR master mix (containing PrimeTime™ Gene Expression Master Mix) at room temperature and still generate consistent results?
The stability of PrimeTime Gene Expression Master Mix makes it ideal for overnight PCR runs and high throughput experiments using robotic liquid handling systems.
Reliable results have been obtained when the qPCR was run directly after setup or after the reactions were maintained at room temperature for up to 72 hr.
Download the white paper, Ambient shipping of PrimeTime Gene Expression Master Mix for more data.
How many freeze-thaw events can Primetime™ Gene Expression Master Mix withstand?
We tested up to 20 freeze-thaw cycles, which is more extreme than typical laboratory usage. No detrimental effects or loss in Cq were observed. However, we still recommend following good laboratory practice by minimizing freeze-thaw cycles.
Download the white paper, Ambient shipping of PrimeTime Gene Expression Master Mix, for more data.