Quick facts
Composition: Chimeric DNA/RNA Oligo
Ordering path: Order as a Custom RNA Oligo
Typical length: ~30 bases
Scales: 100 nmol to large scale synthesis
Purification: Standard desalt; RNase-free HPLC available on request
Website ordering symbol: Use lower case “r” in front to specify ribonucleotide bases (e.g,, ACTGCrGrGrG)
Under-representation of mRNA 5' end sequences
Emerging high-throughput technologies such as RNA-seq have enabled functional dissection of complex transcriptomes with a minimal amount of RNA input. Quantification of individual transcripts or identification of novel transcripts requires an efficient approach to convert mRNA molecules into cDNA. However, conventional cDNA construction methods often result in an under-representation of the 5' end sequences of the mRNA, which poses a technical obstacle [1,2].
SMART technology with a TS Oligo secures 5' end transcript sequences
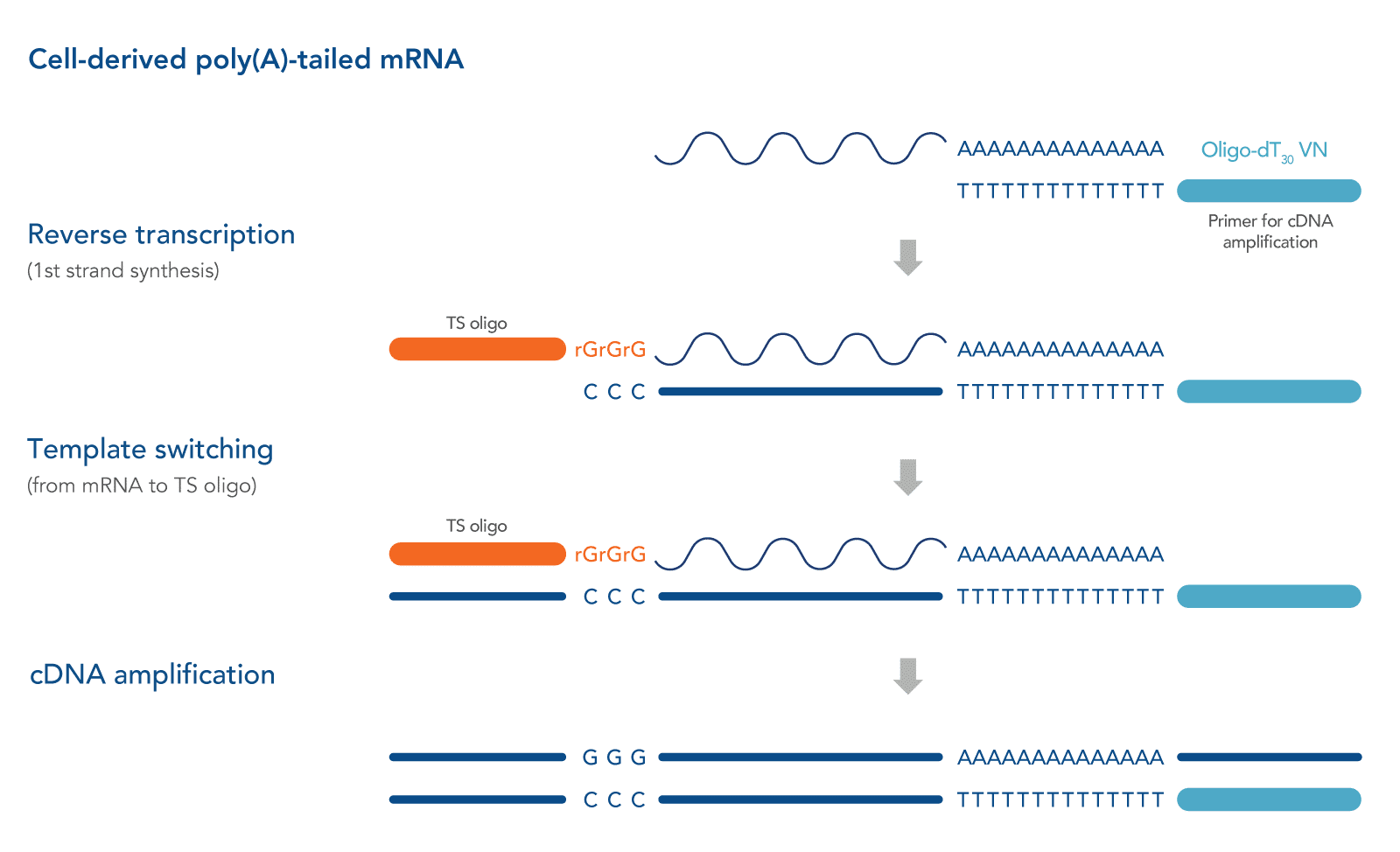
Described originally in 2001, a strategy frequently referred to as “SMART” (switching mechanism at the 5' end of the RNA transcript) technology (Takara Bio USA, Inc) has shown promise in generating full-length cDNA libraries, even from single-cell–derived RNA samples [1,3]. This strategy relies on the intrinsic properties of the Moloney murine leukemia virus (MMLV) reverse transcriptase and the use of a unique TS Oligos. During first-strand synthesis when priming with an oligo dT primer to capture polyadenylated transcripts, upon reaching the 5' end of the RNA template, the terminal transferase activity of the MMLV reverse transcriptase adds a few additional nucleotides (mostly deoxycytidine) to the 3' end of the newly synthesized cDNA strand. These bases function as a TS Oligo-anchoring site. Upon base pairing between the TS Oligo and the appended deoxycytidine stretch, the reverse transcriptase "switches" template strands, from mRNA to the TS Oligo, and continues replication to the 5' end of the TS Oligo. By doing so, the resulting cDNA contains the complete transcript, and universal sequences of choice are added to each end of the reverse transcription product. Along with tagging of the mRNA 3' end by oligo dT primers, we recommend also incorporating a 5’ primer in order amplify the entire full-length cDNA pool in a sequence-independent manner [4]. See Figure 1 for a schematic of this method.
TS Oligo structure
The simple version of a TS Oligo is a DNA oligo sequence that carries 3 riboguanosines (rGrGrG) at its 3' end [1]. The complementarity between these consecutive rG bases and the 3' dC extension of the cDNA molecule empowers the subsequent template switching [5]. A more recent study suggested replacing the 3' most rG with a locked nucleic acid base to enhance thermostability, which is advantageous for the short stretch of base pairing [6].
Interestingly, a systematic study was conducted in 2013 to compare the multiple modified TS Oligo versions side-by-side, and the data indicated that RNA/DNA hybrids provide a superior level of 5' cap-specific enrichment, even though the DNA/locked nucleic acid duplexes were expected to be more thermodynamically favored [7].
Driven by the growing interest in transcriptomics, alternative modification patterns have been explored to further improve TS Oligo performance. For instance, in 2010 researchers reported an approach to reduce library background and thereby improve cDNA yield by incorporating isomeric nucleotides into the TS Oligo [8]. In this study, 2 modified bases, iso-dC and iso-dG, were appended to the 5’ end of the TS Oligo. These 2 modifications, which are chemical variants of cytosine and guanine, respectively, form hydrogen bonds with each other but not with naturally occurring C and G nucleotides. As anticipated, this modified version showed efficacy in minimizing the concatenation of TS Oligos, which typically results from cycles of reverse transcriptase activity [8,9].
Ordering TS Oligos
Taken together, TS Oligos attach a universal primer-binding site through the activity of MMLV reverse transcriptase, enabling exclusive amplification of intact cDNA molecules. TS Oligos containing 3' rGs can be ordered from the Custom RNA Oligos ordering page on the IDT website. You can further customize your TS Oligo with unique properties by incorporating additional chemical modifications from this same ordering page.
Contact us with any questions about ordering modified oligos or to discuss your experimental design.
References
- Zhu YY, Machleder EM, Chenchik A, et al. Reverse transcriptase template switching: a SMART approach for full-length cDNA library construction. Biotechniques. 2001;30(4):892-897.
- Wellenreuther R, Schupp I, Poustka A, et al. SMART amplification combined with cDNA size fractionation in order to obtain large full-length clones. BMC Genomics. 2004;5(1):36.
- Ramskold D, Luo S, Wang YC, et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat Biotechnol. 2012;30(8):777-782.
- Shapiro E, Biezuner T, Linnarsson S. Single-cell sequencing-based technologies will revolutionize whole-organism science. Nat Rev Genet. 2013;14(9):618-630.
- Turchinovich A, Surowy H, Serva A, et al. Capture and Amplification by Tailing and Switching (CATS). An ultrasensitive ligation-independent method for generation of DNA libraries for deep sequencing from picogram amounts of DNA and RNA. RNA Biol. 2014;11(7):817-828.
- Picelli S, Faridani OR, Bjorklund AK, et al. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc. 2014;9(1):171-181.
- Harbers M, Kato S, de Hoon M, et al. Comparison of RNA- or LNA-hybrid oligonucleotides in template-switching reactions for high-speed sequencing library preparation. BMC Genomics. 2013;14:665.
- Kapteyn J, He R, McDowell ET, et al. Incorporation of non-natural nucleotides into template-switching oligonucleotides reduces background and improves cDNA synthesis from very small RNA samples. BMC Genomics. 2010;11:413.
- Saliba AE, Li L, Westermann AJ, et al. Single-cell RNA-seq ties macrophage polarization to growth rate of intracellular Salmonella. Nat Microbiol. 2016;2:16206.
For research use only. Not for use in diagnostic procedures. Unless otherwise agreed to in writing, IDT does not intend these products to be used in clinical applications and does not warrant their fitness or suitability for any clinical diagnostic use. Purchaser is solely responsible for all decisions regarding the use of these products and any associated regulatory or legal obligations. Doc ID: RUO22-1621_001