What is NGS library prep?
Next generation sequencing (NGS) allows researchers to generate millions of reads in a single sequencing run, but before all that data can be generated sequencing libraries have to be prepared. After nucleic acid extraction, the following step of a typical NGS workflow consists of library preparation, or library prep (Figure 1).
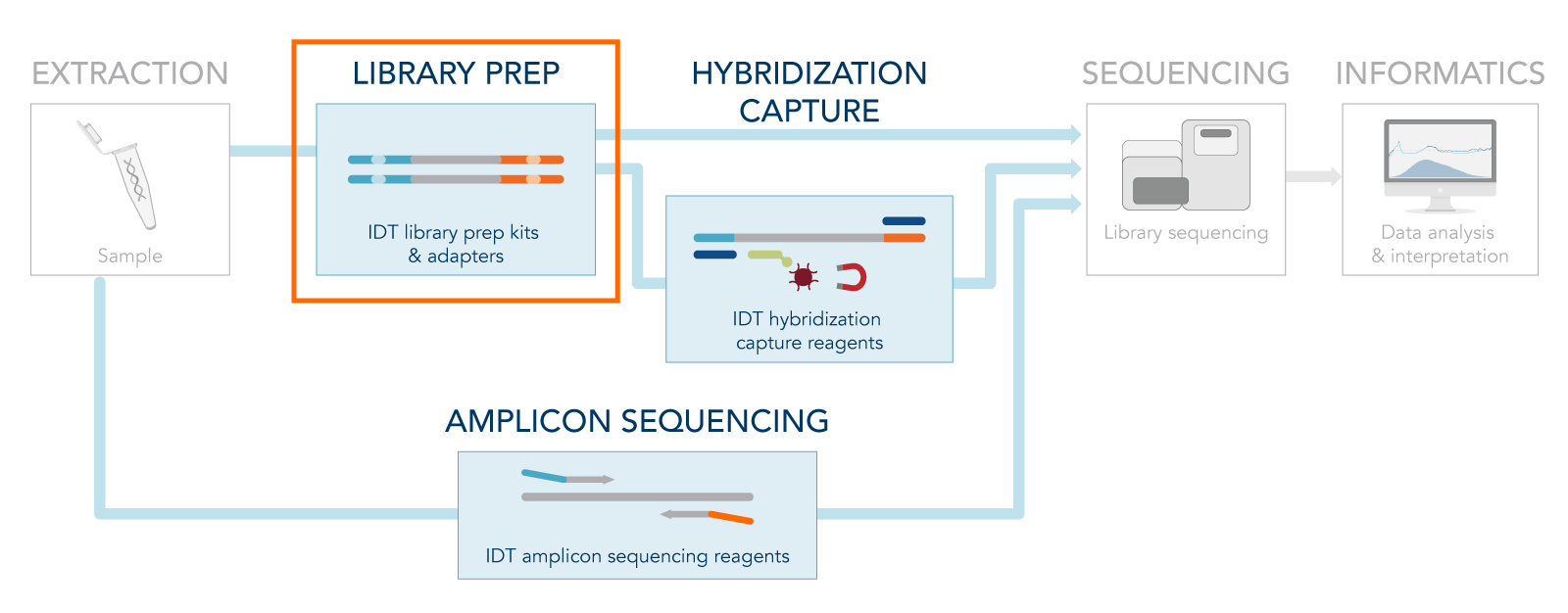
This step prepares DNA or cDNA (DNA synthesized from RNA using a reverse transcriptase) molecules for sequencing. The goal of NGS library prep is to generate a pool of DNA fragments of a similar size that have been labeled with adapter sequences and indexes. Adapters are designed to make DNA molecules compatible with a specific library sequencing platform and indexes aid with downstream sample identification.
How is NGS library prep completed?
The preparation of DNA or cDNA molecules for library sequencing consists of fragmentation, end-repair, adapter addition, and an optional PCR amplification step.
- DNA extracted from a sample will need to undergo either fragmentation or size selection to help ensure that the molecules in your sequencing library are of an appropriate length for your sequencing platform chemistry. Short‑read platforms – e.g., Illumina® – are designed to read DNA molecules of a specific length and are not compatible with long sequences of DNA. The size of library molecules is unique and should be selected with a research objective as well as desired sequencing platform in mind.
- Once DNA/cDNA molecules have been fragmented to an appropriate length, sequences are end repaired. This step prepares DNA molecules for the addition of sequencing adapters.
- Once ends are ready, adapters can either be added directly to the ‘blunt’ end fragments or an A-tail can be added to the fragments to prepare for the addition of adapters. Again, this depends upon which sequencing chemistry will be used. Adapters are specific to sequencing platforms and should be selected carefully. They can also include sequencing indexes or barcodes, which help with the identification of samples during analyses and allow libraries to be multiplexed. After adapters have been added to library molecules, libraries should be quantified, and the size of library molecules should be confirmed.
- An optional PCR amplification step can be used to generate a more concentrated library. The necessity for this step largely depends on research objectives as well as the sequencing platform being used. If libraries are amplified, a final PCR-clean up step should be performed to remove any remaining primer-dimers and small fragments from the final product.
To learn more about specific NGS library prep workflows, download the Next generation sequencing guide here.
Picking the right NGS library prep kit
Knowing which NGS library prep kit to pick can be overwhelming, but IDT is here to help. The free xGen™ NGS Solutions Builder Tool provides library prep kit suggestions tailored to your research objectives.
Simply select the workflow step you’re interested in – e.g., Library Prep and Hyb Capture. Then choose from DNA sequencing (xGen DNA-seq), RNA sequencing (RNA-seq), or methylation sequencing (Methyl-seq); and click Continue to view xGen Library Preparation Kits available from IDT.
To finish, select the additional library prep features you’ll need (i.e., indexing and fragmentation methods, normalization, etc.), and the xGen NGS Solutions Builder Tool will provide a complete list of recommended kits for your workflow.
Click here to get started using the xGen NGS Solutions Builder Tool, or contact us to get additional information about NGS library prep.
RUO23-2301_001